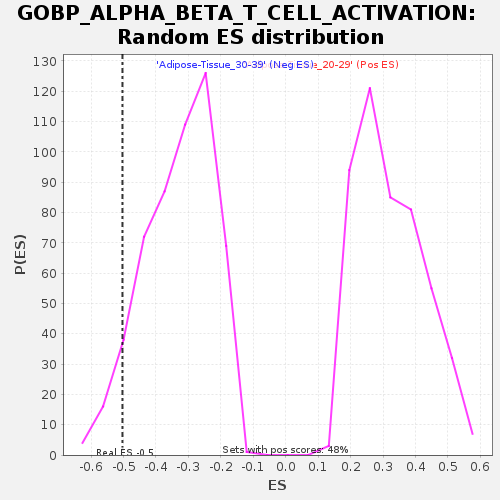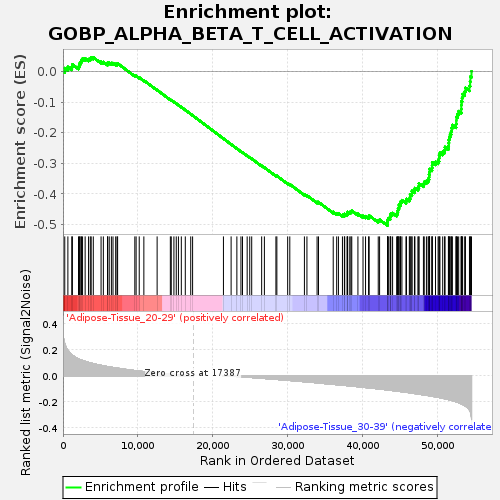
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_30-39 |
| GeneSet | GOBP_ALPHA_BETA_T_CELL_ACTIVATION |
| Enrichment Score (ES) | -0.5032652 |
| Normalized Enrichment Score (NES) | -1.5215048 |
| Nominal p-value | 0.0651341 |
| FDR q-value | 0.84663135 |
| FWER p-Value | 0.949 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | STOML2 | NA | 234 | 0.247 | 0.0105 | No |
| 2 | CTSL | NA | 652 | 0.195 | 0.0145 | No |
| 3 | ENTPD7 | NA | 1172 | 0.162 | 0.0147 | No |
| 4 | STAT3 | NA | 1255 | 0.158 | 0.0226 | No |
| 5 | PRR7 | NA | 2073 | 0.129 | 0.0153 | No |
| 6 | ARG2 | NA | 2185 | 0.126 | 0.0207 | No |
| 7 | AP3D1 | NA | 2245 | 0.125 | 0.0271 | No |
| 8 | PNP | NA | 2395 | 0.121 | 0.0316 | No |
| 9 | BCL3 | NA | 2464 | 0.120 | 0.0375 | No |
| 10 | RELB | NA | 2612 | 0.117 | 0.0418 | No |
| 11 | RUNX1 | NA | 2958 | 0.110 | 0.0421 | No |
| 12 | SOCS1 | NA | 3414 | 0.103 | 0.0399 | No |
| 13 | ABL1 | NA | 3645 | 0.099 | 0.0416 | No |
| 14 | ZFPM1 | NA | 3764 | 0.097 | 0.0452 | No |
| 15 | CD81 | NA | 4048 | 0.093 | 0.0456 | No |
| 16 | SMAD7 | NA | 5086 | 0.080 | 0.0313 | No |
| 17 | ZC3H12A | NA | 5390 | 0.077 | 0.0304 | No |
| 18 | IL12A | NA | 5958 | 0.070 | 0.0241 | No |
| 19 | IL6 | NA | 5959 | 0.070 | 0.0283 | No |
| 20 | SEMA4A | NA | 6175 | 0.068 | 0.0284 | No |
| 21 | GATA3 | NA | 6465 | 0.065 | 0.0270 | No |
| 22 | ZBTB7B | NA | 6681 | 0.063 | 0.0269 | No |
| 23 | CD55 | NA | 7037 | 0.060 | 0.0239 | No |
| 24 | RARA | NA | 7217 | 0.059 | 0.0241 | No |
| 25 | MIR21 | NA | 7297 | 0.058 | 0.0262 | No |
| 26 | HSPH1 | NA | 9596 | 0.040 | -0.0137 | No |
| 27 | ANXA1 | NA | 9769 | 0.039 | -0.0145 | No |
| 28 | NFKBIZ | NA | 10185 | 0.036 | -0.0200 | No |
| 29 | IL23A | NA | 10803 | 0.032 | -0.0295 | No |
| 30 | LEF1 | NA | 12576 | 0.021 | -0.0608 | No |
| 31 | JAK3 | NA | 14342 | 0.012 | -0.0924 | No |
| 32 | CD274 | NA | 14465 | 0.012 | -0.0940 | No |
| 33 | MAPK8IP1 | NA | 14819 | 0.010 | -0.0998 | No |
| 34 | RORC | NA | 15133 | 0.009 | -0.1051 | No |
| 35 | NKX2-3 | NA | 15423 | 0.008 | -0.1099 | No |
| 36 | GLI3 | NA | 15809 | 0.006 | -0.1166 | No |
| 37 | IRF1 | NA | 16340 | 0.004 | -0.1261 | No |
| 38 | TARM1 | NA | 17067 | 0.001 | -0.1394 | No |
| 39 | IL4R | NA | 17317 | 0.000 | -0.1439 | No |
| 40 | FUT7 | NA | 21437 | -0.000 | -0.2195 | No |
| 41 | IL12B | NA | 22462 | -0.004 | -0.2381 | No |
| 42 | CBFB | NA | 23231 | -0.006 | -0.2518 | No |
| 43 | MTOR | NA | 23776 | -0.008 | -0.2614 | No |
| 44 | BCL2 | NA | 23983 | -0.009 | -0.2646 | No |
| 45 | PRKCZ | NA | 24607 | -0.011 | -0.2754 | No |
| 46 | AP3B1 | NA | 24951 | -0.012 | -0.2809 | No |
| 47 | NFKBID | NA | 25213 | -0.013 | -0.2849 | No |
| 48 | CCR2 | NA | 26543 | -0.019 | -0.3082 | No |
| 49 | IL18R1 | NA | 26908 | -0.021 | -0.3136 | No |
| 50 | BATF | NA | 28448 | -0.028 | -0.3402 | No |
| 51 | IHH | NA | 28575 | -0.028 | -0.3409 | No |
| 52 | PTGER4 | NA | 30018 | -0.035 | -0.3653 | No |
| 53 | TMEM98 | NA | 30303 | -0.036 | -0.3683 | No |
| 54 | IL4 | NA | 32256 | -0.046 | -0.4014 | No |
| 55 | ITPKB | NA | 32593 | -0.047 | -0.4048 | No |
| 56 | TNFSF18 | NA | 33948 | -0.054 | -0.4264 | No |
| 57 | IL23R | NA | 34139 | -0.055 | -0.4266 | No |
| 58 | LGALS7B | NA | 36105 | -0.066 | -0.4587 | No |
| 59 | RIPK2 | NA | 36568 | -0.069 | -0.4631 | No |
| 60 | SH3RF1 | NA | 36800 | -0.070 | -0.4632 | No |
| 61 | INS | NA | 37337 | -0.073 | -0.4687 | No |
| 62 | CCL19 | NA | 37597 | -0.074 | -0.4690 | No |
| 63 | HLX | NA | 37639 | -0.074 | -0.4653 | No |
| 64 | VSIR | NA | 37952 | -0.076 | -0.4665 | No |
| 65 | IL6R | NA | 38015 | -0.076 | -0.4631 | No |
| 66 | FOXP3 | NA | 38039 | -0.077 | -0.4589 | No |
| 67 | IL21 | NA | 38317 | -0.078 | -0.4594 | No |
| 68 | NLRP3 | NA | 38469 | -0.079 | -0.4574 | No |
| 69 | RPL22 | NA | 38583 | -0.080 | -0.4547 | No |
| 70 | IRF4 | NA | 39406 | -0.084 | -0.4648 | No |
| 71 | CD86 | NA | 40086 | -0.089 | -0.4720 | No |
| 72 | PLA2G2D | NA | 40425 | -0.091 | -0.4728 | No |
| 73 | ZNF683 | NA | 40831 | -0.093 | -0.4746 | No |
| 74 | ELF4 | NA | 40904 | -0.093 | -0.4704 | No |
| 75 | CLEC4A | NA | 42121 | -0.101 | -0.4867 | No |
| 76 | IL27 | NA | 42301 | -0.102 | -0.4839 | No |
| 77 | SOCS5 | NA | 43359 | -0.109 | -0.4967 | Yes |
| 78 | CD300A | NA | 43361 | -0.109 | -0.4902 | Yes |
| 79 | SHH | NA | 43384 | -0.109 | -0.4841 | Yes |
| 80 | CD83 | NA | 43468 | -0.110 | -0.4790 | Yes |
| 81 | LILRB1 | NA | 43720 | -0.112 | -0.4770 | Yes |
| 82 | TNFSF8 | NA | 43733 | -0.112 | -0.4705 | Yes |
| 83 | CBLB | NA | 43798 | -0.112 | -0.4650 | Yes |
| 84 | PTPRC | NA | 44039 | -0.114 | -0.4626 | Yes |
| 85 | PAX1 | NA | 44583 | -0.118 | -0.4655 | Yes |
| 86 | SASH3 | NA | 44692 | -0.119 | -0.4604 | Yes |
| 87 | TNFSF4 | NA | 44738 | -0.119 | -0.4541 | Yes |
| 88 | BCL6 | NA | 44780 | -0.119 | -0.4477 | Yes |
| 89 | TGFBR2 | NA | 44890 | -0.120 | -0.4425 | Yes |
| 90 | AGER | NA | 44895 | -0.120 | -0.4354 | Yes |
| 91 | LILRB4 | NA | 45096 | -0.122 | -0.4318 | Yes |
| 92 | PRDM1 | NA | 45110 | -0.122 | -0.4247 | Yes |
| 93 | ZBTB16 | NA | 45285 | -0.123 | -0.4205 | Yes |
| 94 | CD28 | NA | 45825 | -0.127 | -0.4228 | Yes |
| 95 | GPR183 | NA | 45898 | -0.128 | -0.4165 | Yes |
| 96 | LGALS9 | NA | 46274 | -0.131 | -0.4156 | Yes |
| 97 | CD3E | NA | 46377 | -0.132 | -0.4096 | Yes |
| 98 | LGALS9C | NA | 46387 | -0.132 | -0.4019 | Yes |
| 99 | IL12RB1 | NA | 46615 | -0.134 | -0.3981 | Yes |
| 100 | ADA | NA | 46625 | -0.134 | -0.3902 | Yes |
| 101 | NKAP | NA | 46960 | -0.137 | -0.3882 | Yes |
| 102 | IL18 | NA | 47019 | -0.137 | -0.3810 | Yes |
| 103 | DOCK2 | NA | 47423 | -0.141 | -0.3800 | Yes |
| 104 | PSMB11 | NA | 47545 | -0.142 | -0.3738 | Yes |
| 105 | RASAL3 | NA | 47558 | -0.142 | -0.3656 | Yes |
| 106 | TWSG1 | NA | 48158 | -0.147 | -0.3678 | Yes |
| 107 | EBI3 | NA | 48264 | -0.148 | -0.3608 | Yes |
| 108 | PTPN22 | NA | 48559 | -0.151 | -0.3572 | Yes |
| 109 | NCKAP1L | NA | 48786 | -0.153 | -0.3523 | Yes |
| 110 | CD80 | NA | 48902 | -0.154 | -0.3452 | Yes |
| 111 | SYK | NA | 48916 | -0.154 | -0.3362 | Yes |
| 112 | WDFY4 | NA | 48969 | -0.155 | -0.3279 | Yes |
| 113 | IL15 | NA | 48982 | -0.155 | -0.3189 | Yes |
| 114 | LOXL3 | NA | 49294 | -0.158 | -0.3152 | Yes |
| 115 | HLA-A | NA | 49322 | -0.158 | -0.3063 | Yes |
| 116 | EOMES | NA | 49334 | -0.158 | -0.2970 | Yes |
| 117 | HMGB1 | NA | 49773 | -0.163 | -0.2953 | Yes |
| 118 | MYB | NA | 50119 | -0.166 | -0.2917 | Yes |
| 119 | CRTAM | NA | 50250 | -0.168 | -0.2841 | Yes |
| 120 | SLAMF6 | NA | 50263 | -0.168 | -0.2743 | Yes |
| 121 | TOX | NA | 50364 | -0.169 | -0.2660 | Yes |
| 122 | CDH26 | NA | 50733 | -0.174 | -0.2624 | Yes |
| 123 | SPN | NA | 50985 | -0.177 | -0.2564 | Yes |
| 124 | XCL1 | NA | 51042 | -0.178 | -0.2468 | Yes |
| 125 | ZAP70 | NA | 51495 | -0.184 | -0.2442 | Yes |
| 126 | BCL11B | NA | 51538 | -0.185 | -0.2339 | Yes |
| 127 | LGALS9B | NA | 51562 | -0.185 | -0.2233 | Yes |
| 128 | RUNX3 | NA | 51629 | -0.186 | -0.2134 | Yes |
| 129 | RC3H2 | NA | 51730 | -0.187 | -0.2040 | Yes |
| 130 | TCIRG1 | NA | 51868 | -0.189 | -0.1952 | Yes |
| 131 | LY9 | NA | 51926 | -0.190 | -0.1849 | Yes |
| 132 | IL2 | NA | 52024 | -0.192 | -0.1752 | Yes |
| 133 | RSAD2 | NA | 52512 | -0.200 | -0.1722 | Yes |
| 134 | IFNG | NA | 52552 | -0.201 | -0.1609 | Yes |
| 135 | RC3H1 | NA | 52555 | -0.201 | -0.1489 | Yes |
| 136 | HLA-DRB1 | NA | 52708 | -0.204 | -0.1395 | Yes |
| 137 | HLA-DRA | NA | 52868 | -0.208 | -0.1300 | Yes |
| 138 | MALT1 | NA | 53209 | -0.216 | -0.1233 | Yes |
| 139 | FOXP1 | NA | 53221 | -0.216 | -0.1106 | Yes |
| 140 | CD160 | NA | 53232 | -0.217 | -0.0978 | Yes |
| 141 | RORA | NA | 53327 | -0.219 | -0.0865 | Yes |
| 142 | ATP7A | NA | 53392 | -0.221 | -0.0744 | Yes |
| 143 | PRKCQ | NA | 53674 | -0.231 | -0.0658 | Yes |
| 144 | HLA-E | NA | 53772 | -0.234 | -0.0536 | Yes |
| 145 | ITK | NA | 54309 | -0.263 | -0.0477 | Yes |
| 146 | TBX21 | NA | 54399 | -0.273 | -0.0330 | Yes |
| 147 | HFE | NA | 54436 | -0.278 | -0.0171 | Yes |
| 148 | TNFRSF14 | NA | 54580 | -0.333 | 0.0002 | Yes |