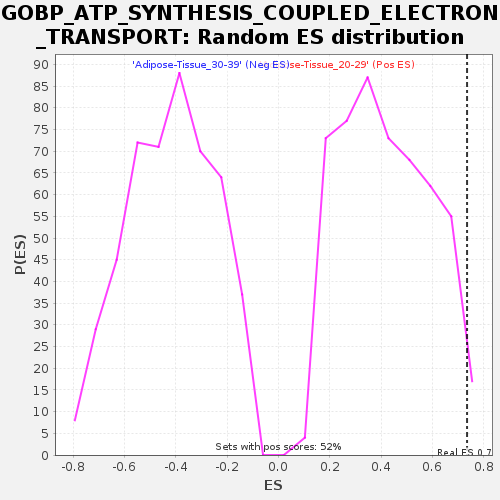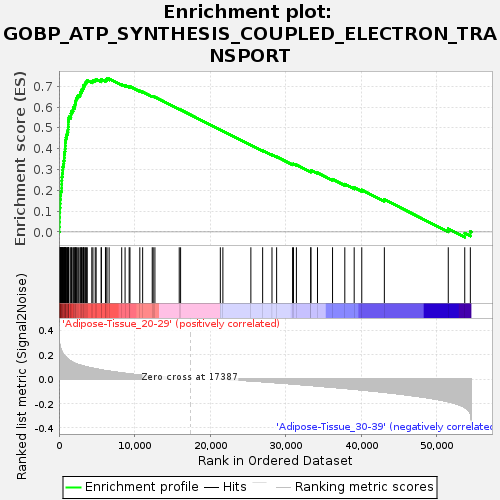
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_20-29 |
| GeneSet | GOBP_ATP_SYNTHESIS_COUPLED_ELECTRON_TRANSPORT |
| Enrichment Score (ES) | 0.7350188 |
| Normalized Enrichment Score (NES) | 1.7507061 |
| Nominal p-value | 0.015503876 |
| FDR q-value | 0.2625251 |
| FWER p-Value | 0.596 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | NDUFS2 | NA | 42 | 0.301 | 0.0234 | Yes |
| 2 | CYC1 | NA | 55 | 0.297 | 0.0470 | Yes |
| 3 | COX10 | NA | 69 | 0.291 | 0.0701 | Yes |
| 4 | NDUFAB1 | NA | 80 | 0.286 | 0.0928 | Yes |
| 5 | UQCRHL | NA | 110 | 0.276 | 0.1144 | Yes |
| 6 | UQCRFS1 | NA | 139 | 0.267 | 0.1352 | Yes |
| 7 | COX5A | NA | 160 | 0.260 | 0.1557 | Yes |
| 8 | UQCRH | NA | 191 | 0.255 | 0.1756 | Yes |
| 9 | UQCRFS1P1 | NA | 233 | 0.247 | 0.1946 | Yes |
| 10 | COX8A | NA | 355 | 0.228 | 0.2107 | Yes |
| 11 | NDUFA8 | NA | 359 | 0.227 | 0.2288 | Yes |
| 12 | NDUFS5 | NA | 370 | 0.224 | 0.2466 | Yes |
| 13 | NDUFA6 | NA | 385 | 0.223 | 0.2642 | Yes |
| 14 | CYCS | NA | 449 | 0.215 | 0.2803 | Yes |
| 15 | UQCRC2 | NA | 481 | 0.211 | 0.2966 | Yes |
| 16 | NDUFA9 | NA | 512 | 0.209 | 0.3128 | Yes |
| 17 | COX7B | NA | 601 | 0.200 | 0.3272 | Yes |
| 18 | DLD | NA | 645 | 0.196 | 0.3421 | Yes |
| 19 | UQCRC1 | NA | 698 | 0.192 | 0.3565 | Yes |
| 20 | COA6 | NA | 714 | 0.191 | 0.3716 | Yes |
| 21 | NDUFS1 | NA | 761 | 0.188 | 0.3858 | Yes |
| 22 | COX7C | NA | 806 | 0.184 | 0.3998 | Yes |
| 23 | NDUFAF1 | NA | 832 | 0.183 | 0.4140 | Yes |
| 24 | NDUFS6 | NA | 842 | 0.182 | 0.4284 | Yes |
| 25 | NDUFC2 | NA | 864 | 0.180 | 0.4424 | Yes |
| 26 | COX5B | NA | 930 | 0.177 | 0.4554 | Yes |
| 27 | COX6A1 | NA | 1008 | 0.172 | 0.4677 | Yes |
| 28 | COX6B1 | NA | 1118 | 0.165 | 0.4789 | Yes |
| 29 | NDUFV2 | NA | 1184 | 0.162 | 0.4907 | Yes |
| 30 | NDUFB9 | NA | 1210 | 0.160 | 0.5031 | Yes |
| 31 | NDUFB6 | NA | 1229 | 0.159 | 0.5155 | Yes |
| 32 | NDUFA1 | NA | 1232 | 0.159 | 0.5282 | Yes |
| 33 | UQCRQ | NA | 1262 | 0.158 | 0.5403 | Yes |
| 34 | NDUFA4 | NA | 1317 | 0.156 | 0.5518 | Yes |
| 35 | PARK7 | NA | 1571 | 0.146 | 0.5588 | Yes |
| 36 | NDUFA7 | NA | 1580 | 0.145 | 0.5703 | Yes |
| 37 | UQCR10 | NA | 1669 | 0.142 | 0.5800 | Yes |
| 38 | COQ9 | NA | 1858 | 0.135 | 0.5874 | Yes |
| 39 | CHCHD2 | NA | 1884 | 0.134 | 0.5978 | Yes |
| 40 | NDUFB2 | NA | 2084 | 0.128 | 0.6044 | Yes |
| 41 | MT-CO1 | NA | 2118 | 0.127 | 0.6140 | Yes |
| 42 | NDUFV3 | NA | 2156 | 0.126 | 0.6235 | Yes |
| 43 | SDHA | NA | 2223 | 0.125 | 0.6323 | Yes |
| 44 | NDUFB3 | NA | 2295 | 0.124 | 0.6409 | Yes |
| 45 | GHITM | NA | 2408 | 0.121 | 0.6485 | Yes |
| 46 | MT-ND6 | NA | 2559 | 0.118 | 0.6552 | Yes |
| 47 | NDUFS3 | NA | 2781 | 0.114 | 0.6603 | Yes |
| 48 | NDUFB8 | NA | 2814 | 0.113 | 0.6688 | Yes |
| 49 | COX6C | NA | 2950 | 0.111 | 0.6752 | Yes |
| 50 | MT-ND5 | NA | 2990 | 0.110 | 0.6833 | Yes |
| 51 | NDUFB11 | NA | 3141 | 0.107 | 0.6891 | Yes |
| 52 | NDUFA13 | NA | 3239 | 0.106 | 0.6958 | Yes |
| 53 | NDUFA12 | NA | 3245 | 0.106 | 0.7042 | Yes |
| 54 | MT-ND4 | NA | 3412 | 0.103 | 0.7094 | Yes |
| 55 | COX4I1 | NA | 3492 | 0.102 | 0.7161 | Yes |
| 56 | MT-ND4L | NA | 3628 | 0.099 | 0.7216 | Yes |
| 57 | NDUFS8 | NA | 3741 | 0.098 | 0.7273 | Yes |
| 58 | UQCC3 | NA | 4344 | 0.089 | 0.7234 | Yes |
| 59 | NDUFS7 | NA | 4492 | 0.087 | 0.7277 | Yes |
| 60 | UQCR11 | NA | 4792 | 0.084 | 0.7289 | Yes |
| 61 | NDUFA10 | NA | 4915 | 0.082 | 0.7333 | Yes |
| 62 | BID | NA | 5583 | 0.074 | 0.7270 | Yes |
| 63 | SDHAF2 | NA | 5610 | 0.074 | 0.7324 | Yes |
| 64 | SDHC | NA | 6151 | 0.068 | 0.7280 | Yes |
| 65 | NDUFA3 | NA | 6211 | 0.068 | 0.7323 | Yes |
| 66 | NDUFB10 | NA | 6353 | 0.066 | 0.7350 | Yes |
| 67 | NDUFB7 | NA | 6634 | 0.064 | 0.7350 | No |
| 68 | NDUFB1 | NA | 8300 | 0.049 | 0.7084 | No |
| 69 | SDHD | NA | 8747 | 0.046 | 0.7039 | No |
| 70 | NDUFA2 | NA | 9287 | 0.042 | 0.6973 | No |
| 71 | CCNB1 | NA | 9393 | 0.041 | 0.6987 | No |
| 72 | NDUFS4 | NA | 10698 | 0.032 | 0.6774 | No |
| 73 | MT-CO2 | NA | 11073 | 0.030 | 0.6729 | No |
| 74 | DNAJC15 | NA | 12342 | 0.022 | 0.6514 | No |
| 75 | MTCO2P12 | NA | 12469 | 0.022 | 0.6508 | No |
| 76 | DGUOK | NA | 12697 | 0.020 | 0.6483 | No |
| 77 | NDUFB4 | NA | 15916 | 0.006 | 0.5897 | No |
| 78 | NDUFB5 | NA | 16097 | 0.005 | 0.5868 | No |
| 79 | UQCRB | NA | 21357 | -0.000 | 0.4903 | No |
| 80 | MT-CO3 | NA | 21694 | -0.002 | 0.4842 | No |
| 81 | NDUFV1 | NA | 25399 | -0.014 | 0.4174 | No |
| 82 | NDUFC1 | NA | 26958 | -0.021 | 0.3905 | No |
| 83 | COX15 | NA | 28194 | -0.027 | 0.3699 | No |
| 84 | NDUFA11 | NA | 28809 | -0.029 | 0.3610 | No |
| 85 | PINK1 | NA | 30921 | -0.039 | 0.3254 | No |
| 86 | CDK1 | NA | 31040 | -0.040 | 0.3265 | No |
| 87 | NDUFA5 | NA | 31427 | -0.042 | 0.3227 | No |
| 88 | MT-CYB | NA | 33330 | -0.051 | 0.2919 | No |
| 89 | MIR210 | NA | 33341 | -0.051 | 0.2958 | No |
| 90 | MT-ND1 | NA | 34219 | -0.056 | 0.2842 | No |
| 91 | COX6A2 | NA | 36212 | -0.067 | 0.2530 | No |
| 92 | COX7A2L | NA | 37835 | -0.075 | 0.2293 | No |
| 93 | SNCA | NA | 39076 | -0.083 | 0.2131 | No |
| 94 | COX4I2 | NA | 40080 | -0.089 | 0.2018 | No |
| 95 | MT-ND3 | NA | 43070 | -0.107 | 0.1556 | No |
| 96 | ISCU | NA | 51531 | -0.184 | 0.0151 | No |
| 97 | MT-ND2 | NA | 53712 | -0.232 | -0.0063 | No |
| 98 | TAZ | NA | 54454 | -0.280 | 0.0025 | No |