Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_30-39 |
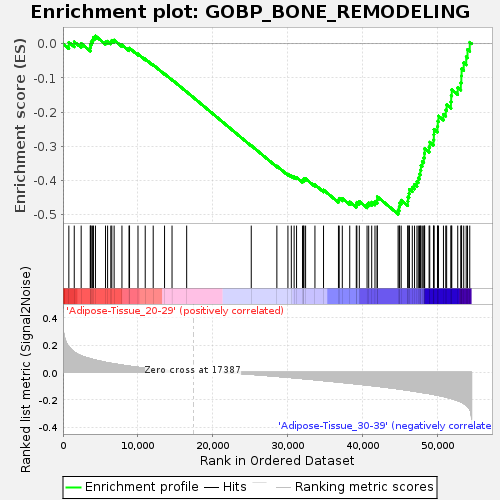
| GeneSet | GOBP_BONE_REMODELING |
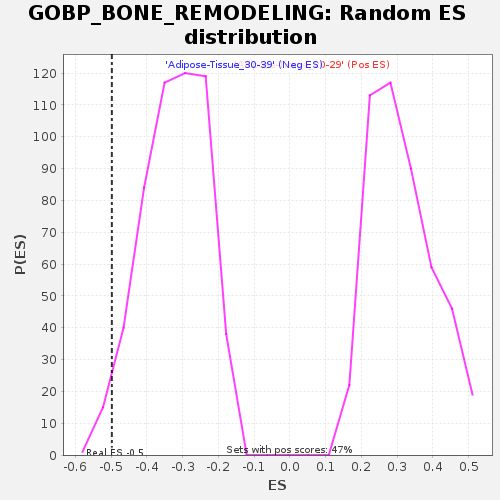
| Enrichment Score (ES) | -0.49789748 |
| Normalized Enrichment Score (NES) | -1.5442256 |
| Nominal p-value | 0.028089888 |
| FDR q-value | 0.88815826 |
| FWER p-Value | 0.941 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | PPARGC1B | NA | 781 | 0.186 | 0.0042 | No |
| 2 | UBASH3B | NA | 1503 | 0.148 | 0.0058 | No |
| 3 | NCDN | NA | 2428 | 0.121 | 0.0009 | No |
| 4 | TNFSF11 | NA | 3647 | 0.099 | -0.0116 | No |
| 5 | ITGB3 | NA | 3666 | 0.099 | -0.0021 | No |
| 6 | GJA1 | NA | 3769 | 0.097 | 0.0058 | No |
| 7 | GDF5 | NA | 3956 | 0.095 | 0.0118 | No |
| 8 | CALCA | NA | 4069 | 0.093 | 0.0190 | No |
| 9 | TFRC | NA | 4336 | 0.089 | 0.0230 | No |
| 10 | GREM1 | NA | 5681 | 0.073 | 0.0056 | No |
| 11 | IL6 | NA | 5959 | 0.070 | 0.0075 | No |
| 12 | MC4R | NA | 6389 | 0.066 | 0.0062 | No |
| 13 | FSHB | NA | 6522 | 0.065 | 0.0102 | No |
| 14 | LGR4 | NA | 6832 | 0.062 | 0.0107 | No |
| 15 | CA2 | NA | 7875 | 0.053 | -0.0031 | No |
| 16 | IAPP | NA | 8822 | 0.045 | -0.0160 | No |
| 17 | SRC | NA | 8890 | 0.045 | -0.0127 | No |
| 18 | TGFB3 | NA | 10019 | 0.037 | -0.0298 | No |
| 19 | ADAM8 | NA | 10986 | 0.030 | -0.0444 | No |
| 20 | P3H4 | NA | 12052 | 0.024 | -0.0616 | No |
| 21 | TNFAIP3 | NA | 13568 | 0.016 | -0.0878 | No |
| 22 | TF | NA | 14568 | 0.011 | -0.1050 | No |
| 23 | CARTPT | NA | 16524 | 0.004 | -0.1405 | No |
| 24 | EPHA2 | NA | 25153 | -0.013 | -0.2975 | No |
| 25 | IHH | NA | 28575 | -0.028 | -0.3574 | No |
| 26 | TNFRSF11B | NA | 30053 | -0.035 | -0.3810 | No |
| 27 | SNX10 | NA | 30506 | -0.037 | -0.3856 | No |
| 28 | ADRB2 | NA | 30882 | -0.039 | -0.3886 | No |
| 29 | MDK | NA | 31207 | -0.041 | -0.3905 | No |
| 30 | CD38 | NA | 31999 | -0.045 | -0.4006 | No |
| 31 | IL20RA | NA | 32149 | -0.045 | -0.3988 | No |
| 32 | CSK | NA | 32187 | -0.046 | -0.3949 | No |
| 33 | S1PR1 | NA | 32415 | -0.047 | -0.3945 | No |
| 34 | PDK4 | NA | 33662 | -0.053 | -0.4121 | No |
| 35 | BGLAP | NA | 34810 | -0.059 | -0.4272 | No |
| 36 | PTN | NA | 36804 | -0.070 | -0.4568 | No |
| 37 | HAMP | NA | 36905 | -0.070 | -0.4516 | No |
| 38 | SUCO | NA | 37319 | -0.073 | -0.4520 | No |
| 39 | LEP | NA | 38312 | -0.078 | -0.4624 | No |
| 40 | SPP2 | NA | 39186 | -0.083 | -0.4701 | No |
| 41 | TMEM64 | NA | 39266 | -0.084 | -0.4633 | No |
| 42 | NOTCH2 | NA | 39593 | -0.086 | -0.4607 | No |
| 43 | PTH | NA | 40626 | -0.092 | -0.4705 | No |
| 44 | EFNA2 | NA | 40846 | -0.093 | -0.4652 | No |
| 45 | WNT16 | NA | 41247 | -0.095 | -0.4631 | No |
| 46 | LRP5 | NA | 41685 | -0.098 | -0.4613 | No |
| 47 | IL7 | NA | 41974 | -0.100 | -0.4566 | No |
| 48 | NF1 | NA | 41991 | -0.100 | -0.4469 | No |
| 49 | LRP6 | NA | 44773 | -0.119 | -0.4860 | Yes |
| 50 | PLEKHM1 | NA | 44919 | -0.121 | -0.4766 | Yes |
| 51 | EGFR | NA | 44979 | -0.121 | -0.4657 | Yes |
| 52 | INPP5D | NA | 45188 | -0.123 | -0.4573 | Yes |
| 53 | TNFRSF11A | NA | 46064 | -0.129 | -0.4605 | Yes |
| 54 | CLDN18 | NA | 46091 | -0.129 | -0.4480 | Yes |
| 55 | CSF1R | NA | 46225 | -0.130 | -0.4375 | Yes |
| 56 | TPP1 | NA | 46288 | -0.131 | -0.4256 | Yes |
| 57 | DEF8 | NA | 46671 | -0.134 | -0.4192 | Yes |
| 58 | HTR1B | NA | 46969 | -0.137 | -0.4110 | Yes |
| 59 | GPR137B | NA | 47294 | -0.139 | -0.4030 | Yes |
| 60 | DCSTAMP | NA | 47471 | -0.141 | -0.3922 | Yes |
| 61 | SPP1 | NA | 47635 | -0.142 | -0.3810 | Yes |
| 62 | RAC1 | NA | 47753 | -0.144 | -0.3689 | Yes |
| 63 | RASSF2 | NA | 47822 | -0.144 | -0.3557 | Yes |
| 64 | TGFB1 | NA | 48001 | -0.146 | -0.3444 | Yes |
| 65 | CTHRC1 | NA | 48204 | -0.148 | -0.3334 | Yes |
| 66 | SIGLEC15 | NA | 48294 | -0.148 | -0.3203 | Yes |
| 67 | LRRK1 | NA | 48323 | -0.149 | -0.3060 | Yes |
| 68 | SYK | NA | 48916 | -0.154 | -0.3015 | Yes |
| 69 | PTK2B | NA | 49003 | -0.155 | -0.2876 | Yes |
| 70 | SYT7 | NA | 49501 | -0.160 | -0.2808 | Yes |
| 71 | RAC2 | NA | 49553 | -0.160 | -0.2657 | Yes |
| 72 | TPH1 | NA | 49568 | -0.161 | -0.2500 | Yes |
| 73 | PRKCA | NA | 50036 | -0.165 | -0.2421 | Yes |
| 74 | GPR137 | NA | 50076 | -0.166 | -0.2263 | Yes |
| 75 | GPR55 | NA | 50167 | -0.167 | -0.2113 | Yes |
| 76 | ACP5 | NA | 50834 | -0.175 | -0.2061 | Yes |
| 77 | LTBP3 | NA | 51143 | -0.179 | -0.1939 | Yes |
| 78 | SFRP1 | NA | 51261 | -0.180 | -0.1781 | Yes |
| 79 | P2RX7 | NA | 51834 | -0.189 | -0.1698 | Yes |
| 80 | TCIRG1 | NA | 51868 | -0.189 | -0.1515 | Yes |
| 81 | LRP5L | NA | 51946 | -0.191 | -0.1339 | Yes |
| 82 | ATP6AP1 | NA | 52741 | -0.205 | -0.1281 | Yes |
| 83 | RAB3D | NA | 53154 | -0.215 | -0.1142 | Yes |
| 84 | MITF | NA | 53223 | -0.216 | -0.0939 | Yes |
| 85 | ZNF675 | NA | 53258 | -0.217 | -0.0729 | Yes |
| 86 | LEPR | NA | 53544 | -0.226 | -0.0556 | Yes |
| 87 | PTH1R | NA | 53875 | -0.237 | -0.0380 | Yes |
| 88 | TRAF6 | NA | 54049 | -0.246 | -0.0167 | Yes |
| 89 | TMEM119 | NA | 54348 | -0.267 | 0.0045 | Yes |