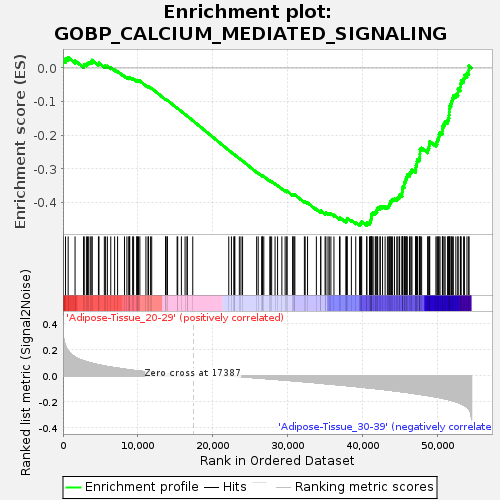
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_30-39 |
| GeneSet | GOBP_CALCIUM_MEDIATED_SIGNALING |
| Enrichment Score (ES) | -0.46865302 |
| Normalized Enrichment Score (NES) | -1.5740997 |
| Nominal p-value | 0.013282732 |
| FDR q-value | 0.9597809 |
| FWER p-Value | 0.921 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | IGF1 | NA | 1 | 0.430 | 0.0222 | No |
| 2 | CIB1 | NA | 338 | 0.231 | 0.0279 | No |
| 3 | CHP1 | NA | 689 | 0.193 | 0.0314 | No |
| 4 | PRKACA | NA | 1611 | 0.144 | 0.0219 | No |
| 5 | BCAP31 | NA | 2749 | 0.115 | 0.0069 | No |
| 6 | ATP2A2 | NA | 2835 | 0.113 | 0.0112 | No |
| 7 | PTBP1 | NA | 3126 | 0.108 | 0.0114 | No |
| 8 | AZU1 | NA | 3277 | 0.105 | 0.0141 | No |
| 9 | P2RX3 | NA | 3430 | 0.103 | 0.0166 | No |
| 10 | TNFSF11 | NA | 3647 | 0.099 | 0.0177 | No |
| 11 | PDK2 | NA | 3874 | 0.096 | 0.0185 | No |
| 12 | VCAM1 | NA | 3876 | 0.096 | 0.0234 | No |
| 13 | CXCL8 | NA | 4736 | 0.084 | 0.0120 | No |
| 14 | GPR143 | NA | 4808 | 0.083 | 0.0150 | No |
| 15 | SLC9A1 | NA | 5524 | 0.075 | 0.0057 | No |
| 16 | SPHK1 | NA | 5694 | 0.073 | 0.0063 | No |
| 17 | LMCD1 | NA | 5931 | 0.071 | 0.0056 | No |
| 18 | ADGRB2 | NA | 6382 | 0.066 | 0.0008 | No |
| 19 | SELE | NA | 6899 | 0.061 | -0.0056 | No |
| 20 | P2RY11 | NA | 7293 | 0.058 | -0.0098 | No |
| 21 | IRGM | NA | 8216 | 0.050 | -0.0242 | No |
| 22 | RCAN1 | NA | 8540 | 0.047 | -0.0276 | No |
| 23 | GRIN2B | NA | 8788 | 0.046 | -0.0298 | No |
| 24 | CCL20 | NA | 8859 | 0.045 | -0.0288 | No |
| 25 | HPCA | NA | 8918 | 0.045 | -0.0276 | No |
| 26 | GRIN1 | NA | 9302 | 0.042 | -0.0325 | No |
| 27 | SAMD14 | NA | 9431 | 0.041 | -0.0327 | No |
| 28 | GRM5 | NA | 9844 | 0.038 | -0.0383 | No |
| 29 | ADGRL1 | NA | 9916 | 0.038 | -0.0377 | No |
| 30 | RIT2 | NA | 9959 | 0.037 | -0.0365 | No |
| 31 | CHERP | NA | 10093 | 0.036 | -0.0371 | No |
| 32 | EDNRB | NA | 10268 | 0.035 | -0.0385 | No |
| 33 | NFATC2 | NA | 11079 | 0.030 | -0.0518 | No |
| 34 | SLC8A2 | NA | 11354 | 0.028 | -0.0554 | No |
| 35 | LACRT | NA | 11431 | 0.028 | -0.0554 | No |
| 36 | FIS1 | NA | 11703 | 0.026 | -0.0590 | No |
| 37 | CCR7 | NA | 11853 | 0.025 | -0.0605 | No |
| 38 | ACKR3 | NA | 13696 | 0.015 | -0.0935 | No |
| 39 | CHP2 | NA | 13736 | 0.015 | -0.0935 | No |
| 40 | ZMPSTE24 | NA | 13893 | 0.014 | -0.0956 | No |
| 41 | MCTP2 | NA | 13951 | 0.014 | -0.0959 | No |
| 42 | CCR1 | NA | 15248 | 0.008 | -0.1193 | No |
| 43 | PLCE1 | NA | 15327 | 0.008 | -0.1203 | No |
| 44 | NFATC1 | NA | 15817 | 0.006 | -0.1290 | No |
| 45 | TNNI3 | NA | 16311 | 0.004 | -0.1379 | No |
| 46 | NEUROD2 | NA | 16574 | 0.003 | -0.1425 | No |
| 47 | PRNP | NA | 16604 | 0.003 | -0.1429 | No |
| 48 | GSK3B | NA | 17338 | 0.000 | -0.1563 | No |
| 49 | DEFB1 | NA | 22147 | -0.003 | -0.2446 | No |
| 50 | ATP1A2 | NA | 22504 | -0.004 | -0.2509 | No |
| 51 | ANK2 | NA | 22821 | -0.005 | -0.2565 | No |
| 52 | FPR2 | NA | 22929 | -0.005 | -0.2582 | No |
| 53 | CXCR1 | NA | 22953 | -0.005 | -0.2583 | No |
| 54 | CCR9 | NA | 23560 | -0.007 | -0.2691 | No |
| 55 | MTOR | NA | 23776 | -0.008 | -0.2726 | No |
| 56 | EDN1 | NA | 23993 | -0.009 | -0.2761 | No |
| 57 | ATP2B4 | NA | 25874 | -0.016 | -0.3098 | No |
| 58 | MIR1-1 | NA | 26082 | -0.017 | -0.3128 | No |
| 59 | CCR2 | NA | 26543 | -0.019 | -0.3202 | No |
| 60 | GRIN2A | NA | 26638 | -0.020 | -0.3209 | No |
| 61 | NMUR2 | NA | 26676 | -0.020 | -0.3206 | No |
| 62 | ACKR4 | NA | 26830 | -0.020 | -0.3224 | No |
| 63 | MAPT | NA | 27641 | -0.024 | -0.3360 | No |
| 64 | CXCR6 | NA | 27707 | -0.024 | -0.3360 | No |
| 65 | CCR3 | NA | 27880 | -0.025 | -0.3378 | No |
| 66 | CXCR2 | NA | 28335 | -0.027 | -0.3448 | No |
| 67 | ERBB3 | NA | 28670 | -0.029 | -0.3494 | No |
| 68 | AVPR1A | NA | 29244 | -0.031 | -0.3583 | No |
| 69 | AGTR1 | NA | 29681 | -0.034 | -0.3646 | No |
| 70 | KDR | NA | 29904 | -0.035 | -0.3669 | No |
| 71 | P2RY12 | NA | 29937 | -0.035 | -0.3657 | No |
| 72 | EIF2AK3 | NA | 30680 | -0.038 | -0.3774 | No |
| 73 | MCTP1 | NA | 30753 | -0.038 | -0.3767 | No |
| 74 | CALM1 | NA | 30883 | -0.039 | -0.3771 | No |
| 75 | DMTN | NA | 30980 | -0.039 | -0.3768 | No |
| 76 | CAMK2D | NA | 32252 | -0.046 | -0.3978 | No |
| 77 | ATP1B1 | NA | 32355 | -0.046 | -0.3973 | No |
| 78 | XCR1 | NA | 32676 | -0.048 | -0.4007 | No |
| 79 | MYOZ2 | NA | 33853 | -0.054 | -0.4196 | No |
| 80 | CCR5 | NA | 34428 | -0.057 | -0.4272 | No |
| 81 | HDAC4 | NA | 34445 | -0.057 | -0.4245 | No |
| 82 | PPP3R1 | NA | 35018 | -0.060 | -0.4319 | No |
| 83 | CXCR3 | NA | 35125 | -0.061 | -0.4307 | No |
| 84 | SELENOK | NA | 35407 | -0.062 | -0.4327 | No |
| 85 | GSTO1 | NA | 35576 | -0.063 | -0.4325 | No |
| 86 | PPP1R9A | NA | 35793 | -0.064 | -0.4331 | No |
| 87 | NUDT4 | NA | 36202 | -0.067 | -0.4372 | No |
| 88 | TNF | NA | 36942 | -0.071 | -0.4472 | No |
| 89 | EFHB | NA | 37017 | -0.071 | -0.4449 | No |
| 90 | KSR2 | NA | 37794 | -0.075 | -0.4552 | No |
| 91 | TRPM5 | NA | 37873 | -0.075 | -0.4528 | No |
| 92 | CCR8 | NA | 37936 | -0.076 | -0.4500 | No |
| 93 | CAMTA1 | NA | 37970 | -0.076 | -0.4467 | No |
| 94 | CALM2 | NA | 38531 | -0.079 | -0.4529 | No |
| 95 | PPP1R9B | NA | 39115 | -0.083 | -0.4594 | No |
| 96 | TPCN2 | NA | 39622 | -0.086 | -0.4642 | Yes |
| 97 | PLEK | NA | 39744 | -0.087 | -0.4620 | Yes |
| 98 | CCL4 | NA | 39842 | -0.087 | -0.4593 | Yes |
| 99 | NMUR1 | NA | 39912 | -0.088 | -0.4560 | Yes |
| 100 | P2RX2 | NA | 40583 | -0.092 | -0.4636 | Yes |
| 101 | GRIN2C | NA | 40593 | -0.092 | -0.4591 | Yes |
| 102 | CHRM3 | NA | 40950 | -0.094 | -0.4608 | Yes |
| 103 | CX3CR1 | NA | 41026 | -0.094 | -0.4573 | Yes |
| 104 | CCR10 | NA | 41120 | -0.095 | -0.4541 | Yes |
| 105 | NFATC3 | NA | 41143 | -0.095 | -0.4496 | Yes |
| 106 | NFATC4 | NA | 41191 | -0.095 | -0.4456 | Yes |
| 107 | TMEM100 | NA | 41244 | -0.095 | -0.4416 | Yes |
| 108 | CAMKK2 | NA | 41257 | -0.095 | -0.4369 | Yes |
| 109 | CCRL2 | NA | 41282 | -0.096 | -0.4324 | Yes |
| 110 | CCL3 | NA | 41436 | -0.097 | -0.4302 | Yes |
| 111 | CALM3 | NA | 41720 | -0.099 | -0.4304 | Yes |
| 112 | C10orf71 | NA | 41797 | -0.099 | -0.4266 | Yes |
| 113 | NR5A1 | NA | 41941 | -0.100 | -0.4241 | Yes |
| 114 | SLC8A1 | NA | 41960 | -0.100 | -0.4193 | Yes |
| 115 | CXCR5 | NA | 42049 | -0.101 | -0.4157 | Yes |
| 116 | ACKR2 | NA | 42351 | -0.103 | -0.4159 | Yes |
| 117 | ACTN3 | NA | 42391 | -0.103 | -0.4113 | Yes |
| 118 | SLA2 | NA | 42698 | -0.105 | -0.4116 | Yes |
| 119 | LAT2 | NA | 43037 | -0.107 | -0.4122 | Yes |
| 120 | TRPM8 | NA | 43344 | -0.109 | -0.4122 | Yes |
| 121 | P2RX5 | NA | 43524 | -0.110 | -0.4098 | Yes |
| 122 | GRIN2D | NA | 43594 | -0.111 | -0.4054 | Yes |
| 123 | BHLHA15 | NA | 43693 | -0.112 | -0.4014 | Yes |
| 124 | HOMER2 | NA | 43725 | -0.112 | -0.3962 | Yes |
| 125 | HTR2B | NA | 43899 | -0.113 | -0.3936 | Yes |
| 126 | PTPRC | NA | 44039 | -0.114 | -0.3902 | Yes |
| 127 | FHL2 | NA | 44289 | -0.116 | -0.3888 | Yes |
| 128 | CMKLR1 | NA | 44597 | -0.118 | -0.3884 | Yes |
| 129 | CDH13 | NA | 44737 | -0.119 | -0.3848 | Yes |
| 130 | EDN2 | NA | 44933 | -0.121 | -0.3821 | Yes |
| 131 | EGFR | NA | 44979 | -0.121 | -0.3767 | Yes |
| 132 | MCOLN1 | NA | 45313 | -0.123 | -0.3765 | Yes |
| 133 | SELP | NA | 45315 | -0.123 | -0.3701 | Yes |
| 134 | PPP3CA | NA | 45336 | -0.124 | -0.3641 | Yes |
| 135 | CCR6 | NA | 45357 | -0.124 | -0.3581 | Yes |
| 136 | TMEM38A | NA | 45403 | -0.124 | -0.3525 | Yes |
| 137 | CASQ2 | NA | 45610 | -0.126 | -0.3498 | Yes |
| 138 | HOMER3 | NA | 45611 | -0.126 | -0.3433 | Yes |
| 139 | PDPK1 | NA | 45667 | -0.126 | -0.3378 | Yes |
| 140 | CD4 | NA | 45788 | -0.127 | -0.3335 | Yes |
| 141 | TMBIM4 | NA | 45843 | -0.127 | -0.3279 | Yes |
| 142 | PPP3CC | NA | 45934 | -0.128 | -0.3229 | Yes |
| 143 | RGN | NA | 45995 | -0.129 | -0.3174 | Yes |
| 144 | PLCG1 | NA | 46292 | -0.131 | -0.3161 | Yes |
| 145 | CD3E | NA | 46377 | -0.132 | -0.3108 | Yes |
| 146 | TREM2 | NA | 46519 | -0.133 | -0.3066 | Yes |
| 147 | ADA | NA | 46625 | -0.134 | -0.3016 | Yes |
| 148 | BST1 | NA | 47114 | -0.138 | -0.3034 | Yes |
| 149 | SULT1A3 | NA | 47143 | -0.138 | -0.2968 | Yes |
| 150 | RYR2 | NA | 47165 | -0.138 | -0.2901 | Yes |
| 151 | TMEM38B | NA | 47270 | -0.139 | -0.2848 | Yes |
| 152 | TBC1D10C | NA | 47279 | -0.139 | -0.2777 | Yes |
| 153 | CCR4 | NA | 47353 | -0.140 | -0.2719 | Yes |
| 154 | DMD | NA | 47602 | -0.142 | -0.2691 | Yes |
| 155 | PTGDR2 | NA | 47679 | -0.143 | -0.2631 | Yes |
| 156 | HTT | NA | 47683 | -0.143 | -0.2558 | Yes |
| 157 | PTGFR | NA | 47696 | -0.143 | -0.2486 | Yes |
| 158 | PRKAA1 | NA | 47699 | -0.143 | -0.2413 | Yes |
| 159 | PTPRJ | NA | 47919 | -0.145 | -0.2378 | Yes |
| 160 | ITPR1 | NA | 48720 | -0.152 | -0.2447 | Yes |
| 161 | PPP3CB | NA | 48795 | -0.153 | -0.2382 | Yes |
| 162 | SYK | NA | 48916 | -0.154 | -0.2324 | Yes |
| 163 | CASQ1 | NA | 48944 | -0.154 | -0.2250 | Yes |
| 164 | PTK2B | NA | 49003 | -0.155 | -0.2180 | Yes |
| 165 | MCU | NA | 49828 | -0.163 | -0.2248 | Yes |
| 166 | AKAP6 | NA | 49969 | -0.165 | -0.2189 | Yes |
| 167 | MAPK7 | NA | 50064 | -0.166 | -0.2120 | Yes |
| 168 | DYRK2 | NA | 50199 | -0.167 | -0.2059 | Yes |
| 169 | NR5A2 | NA | 50230 | -0.168 | -0.1978 | Yes |
| 170 | GBP1 | NA | 50377 | -0.169 | -0.1917 | Yes |
| 171 | TRPM2 | NA | 50695 | -0.173 | -0.1886 | Yes |
| 172 | CLIC2 | NA | 50704 | -0.173 | -0.1798 | Yes |
| 173 | RCAN2 | NA | 50763 | -0.174 | -0.1719 | Yes |
| 174 | P2RX4 | NA | 50913 | -0.176 | -0.1656 | Yes |
| 175 | MYOZ1 | NA | 51058 | -0.178 | -0.1591 | Yes |
| 176 | FKBP1B | NA | 51382 | -0.182 | -0.1556 | Yes |
| 177 | ZAP70 | NA | 51495 | -0.184 | -0.1482 | Yes |
| 178 | CXCR4 | NA | 51573 | -0.185 | -0.1400 | Yes |
| 179 | PLA2G4B | NA | 51604 | -0.185 | -0.1310 | Yes |
| 180 | CACNA1C | NA | 51611 | -0.185 | -0.1216 | Yes |
| 181 | CD22 | NA | 51670 | -0.186 | -0.1130 | Yes |
| 182 | P2RX7 | NA | 51834 | -0.189 | -0.1063 | Yes |
| 183 | PLN | NA | 51923 | -0.190 | -0.0981 | Yes |
| 184 | NFAT5 | NA | 52022 | -0.192 | -0.0900 | Yes |
| 185 | RCAN3 | NA | 52153 | -0.194 | -0.0824 | Yes |
| 186 | BTK | NA | 52490 | -0.200 | -0.0782 | Yes |
| 187 | SPPL3 | NA | 52768 | -0.205 | -0.0727 | Yes |
| 188 | GSTM2 | NA | 52778 | -0.206 | -0.0623 | Yes |
| 189 | TRPM4 | NA | 53090 | -0.213 | -0.0570 | Yes |
| 190 | DMPK | NA | 53110 | -0.214 | -0.0463 | Yes |
| 191 | LRRK2 | NA | 53203 | -0.216 | -0.0369 | Yes |
| 192 | SULT1A4 | NA | 53528 | -0.225 | -0.0312 | Yes |
| 193 | TRAT1 | NA | 53635 | -0.229 | -0.0214 | Yes |
| 194 | PLCG2 | NA | 53975 | -0.242 | -0.0151 | Yes |
| 195 | LAT | NA | 54223 | -0.255 | -0.0065 | Yes |
| 196 | HRC | NA | 54236 | -0.256 | 0.0065 | Yes |