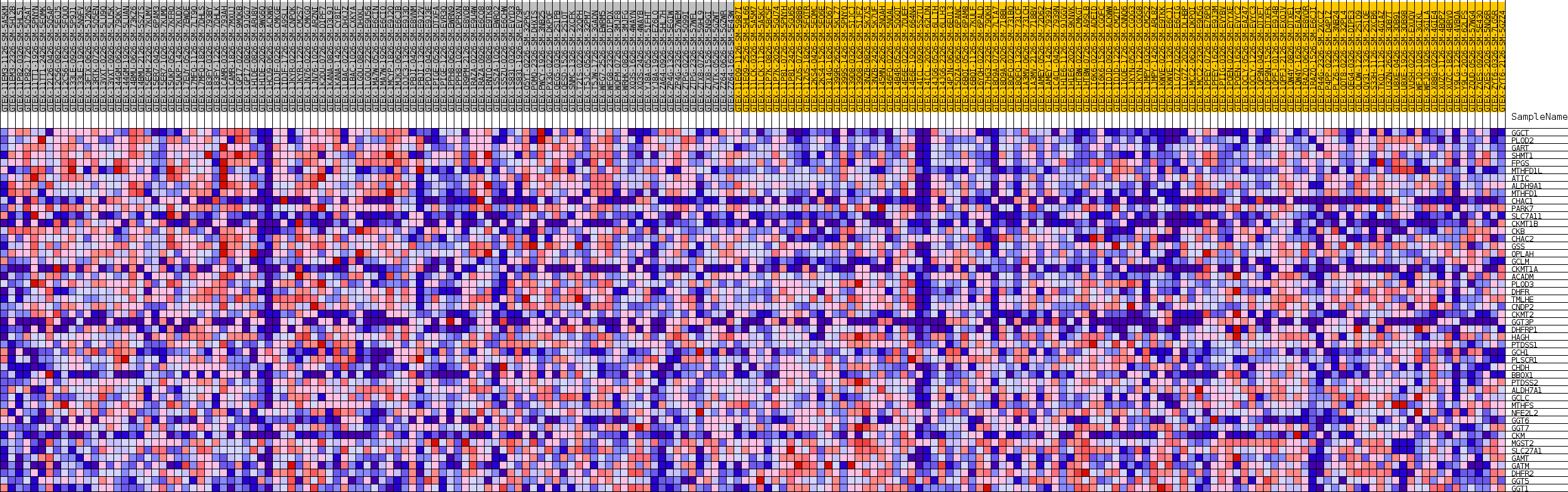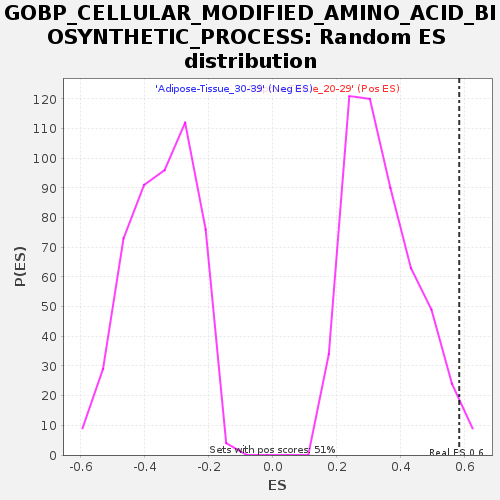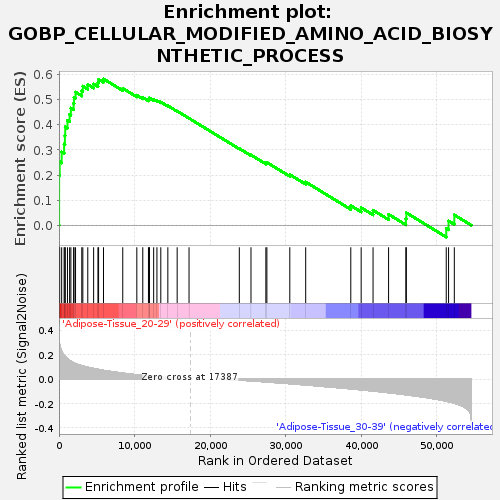
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_20-29 |
| GeneSet | GOBP_CELLULAR_MODIFIED_AMINO_ACID_BIOSYNTHETIC_PROCESS |
| Enrichment Score (ES) | 0.5827803 |
| Normalized Enrichment Score (NES) | 1.7001979 |
| Nominal p-value | 0.021568628 |
| FDR q-value | 0.21342155 |
| FWER p-Value | 0.725 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | GGCT | NA | 13 | 0.351 | 0.0671 | Yes |
| 2 | PLOD2 | NA | 14 | 0.350 | 0.1343 | Yes |
| 3 | GART | NA | 15 | 0.345 | 0.2004 | Yes |
| 4 | SHMT1 | NA | 92 | 0.282 | 0.2531 | Yes |
| 5 | FPGS | NA | 353 | 0.228 | 0.2921 | Yes |
| 6 | MTHFD1L | NA | 677 | 0.194 | 0.3233 | Yes |
| 7 | ATIC | NA | 782 | 0.186 | 0.3572 | Yes |
| 8 | ALDH9A1 | NA | 822 | 0.183 | 0.3916 | Yes |
| 9 | MTHFD1 | NA | 1119 | 0.164 | 0.4178 | Yes |
| 10 | CHAC1 | NA | 1404 | 0.152 | 0.4417 | Yes |
| 11 | PARK7 | NA | 1571 | 0.146 | 0.4666 | Yes |
| 12 | SLC7A11 | NA | 1933 | 0.133 | 0.4855 | Yes |
| 13 | CKMT1B | NA | 1989 | 0.131 | 0.5096 | Yes |
| 14 | CKB | NA | 2189 | 0.126 | 0.5301 | Yes |
| 15 | CHAC2 | NA | 3015 | 0.109 | 0.5359 | Yes |
| 16 | GSS | NA | 3154 | 0.107 | 0.5539 | Yes |
| 17 | OPLAH | NA | 3810 | 0.097 | 0.5605 | Yes |
| 18 | GCLM | NA | 4584 | 0.086 | 0.5629 | Yes |
| 19 | CKMT1A | NA | 5157 | 0.079 | 0.5676 | Yes |
| 20 | ACADM | NA | 5240 | 0.078 | 0.5811 | Yes |
| 21 | PLOD3 | NA | 5890 | 0.071 | 0.5828 | Yes |
| 22 | DHFR | NA | 8439 | 0.048 | 0.5453 | No |
| 23 | TMLHE | NA | 10302 | 0.035 | 0.5179 | No |
| 24 | CNDP2 | NA | 11090 | 0.030 | 0.5092 | No |
| 25 | CKMT2 | NA | 11841 | 0.025 | 0.5003 | No |
| 26 | GGT3P | NA | 11951 | 0.025 | 0.5030 | No |
| 27 | DHFRP1 | NA | 11965 | 0.025 | 0.5075 | No |
| 28 | HAGH | NA | 12531 | 0.021 | 0.5012 | No |
| 29 | PTDSS1 | NA | 12963 | 0.019 | 0.4969 | No |
| 30 | GCH1 | NA | 13475 | 0.016 | 0.4907 | No |
| 31 | PLSCR1 | NA | 14408 | 0.012 | 0.4759 | No |
| 32 | CHDH | NA | 15643 | 0.007 | 0.4546 | No |
| 33 | BBOX1 | NA | 17214 | 0.001 | 0.4260 | No |
| 34 | PTDSS2 | NA | 23870 | -0.008 | 0.3056 | No |
| 35 | ALDH7A1 | NA | 25414 | -0.014 | 0.2800 | No |
| 36 | GCLC | NA | 27384 | -0.023 | 0.2483 | No |
| 37 | MTHFS | NA | 27518 | -0.023 | 0.2504 | No |
| 38 | NFE2L2 | NA | 30552 | -0.037 | 0.2020 | No |
| 39 | GGT6 | NA | 32657 | -0.048 | 0.1726 | No |
| 40 | GGT7 | NA | 38627 | -0.080 | 0.0785 | No |
| 41 | CKM | NA | 40001 | -0.088 | 0.0702 | No |
| 42 | MGST2 | NA | 41574 | -0.098 | 0.0601 | No |
| 43 | SLC27A1 | NA | 43630 | -0.111 | 0.0437 | No |
| 44 | GAMT | NA | 45916 | -0.128 | 0.0264 | No |
| 45 | GATM | NA | 45981 | -0.129 | 0.0499 | No |
| 46 | DHFR2 | NA | 51263 | -0.181 | -0.0123 | No |
| 47 | GGT5 | NA | 51570 | -0.185 | 0.0176 | No |
| 48 | GGT1 | NA | 52331 | -0.197 | 0.0414 | No |