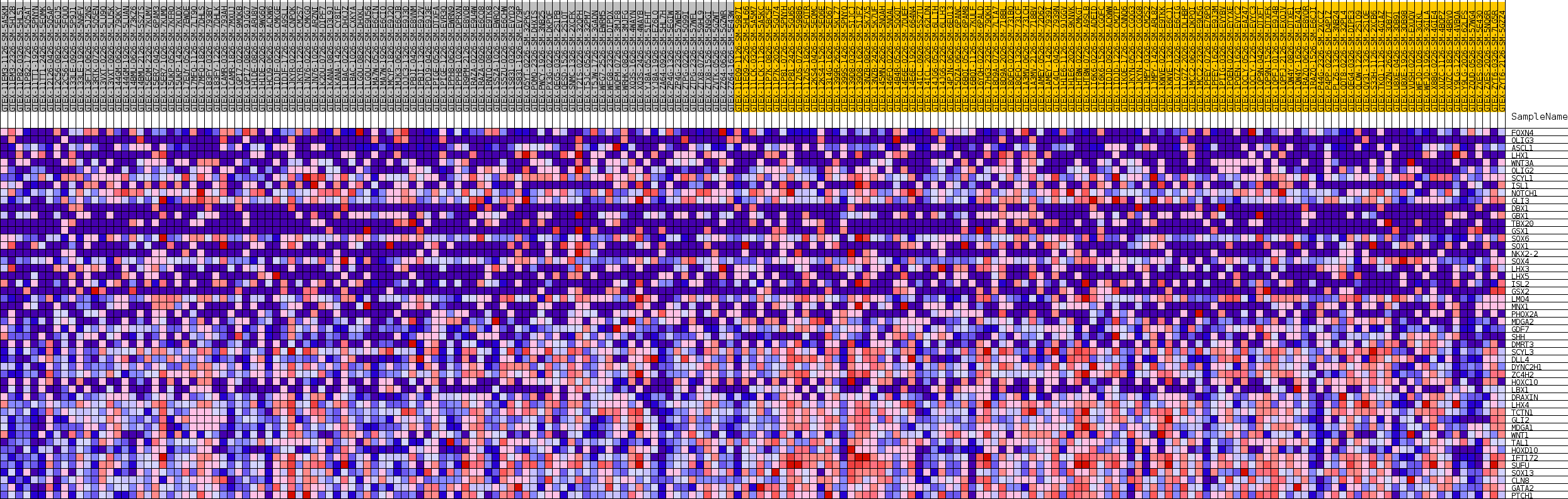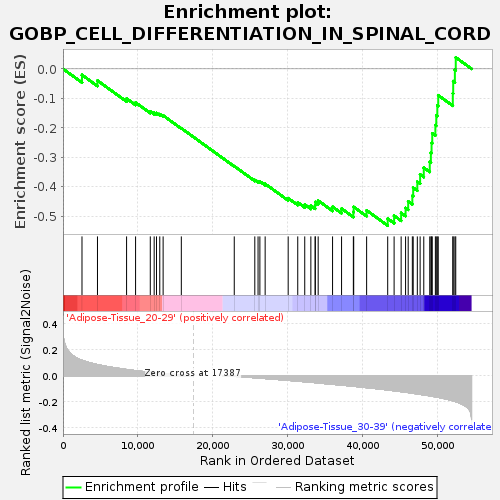
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_30-39 |
| GeneSet | GOBP_CELL_DIFFERENTIATION_IN_SPINAL_CORD |
| Enrichment Score (ES) | -0.53141904 |
| Normalized Enrichment Score (NES) | -1.5970211 |
| Nominal p-value | 0.009881423 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.892 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | FOXN4 | NA | 2537 | 0.119 | -0.0206 | No |
| 2 | OLIG3 | NA | 4606 | 0.086 | -0.0398 | No |
| 3 | ASCL1 | NA | 8495 | 0.048 | -0.1006 | No |
| 4 | LHX1 | NA | 9697 | 0.039 | -0.1141 | No |
| 5 | WNT3A | NA | 11663 | 0.026 | -0.1444 | No |
| 6 | OLIG2 | NA | 12181 | 0.023 | -0.1488 | No |
| 7 | SCYL1 | NA | 12495 | 0.021 | -0.1498 | No |
| 8 | ISL1 | NA | 12930 | 0.019 | -0.1536 | No |
| 9 | NOTCH1 | NA | 13378 | 0.017 | -0.1581 | No |
| 10 | GLI3 | NA | 15809 | 0.006 | -0.2013 | No |
| 11 | DBX1 | NA | 22882 | -0.005 | -0.3299 | No |
| 12 | GBX1 | NA | 25627 | -0.015 | -0.3769 | No |
| 13 | TBX20 | NA | 26062 | -0.017 | -0.3811 | No |
| 14 | GSX1 | NA | 26306 | -0.018 | -0.3816 | No |
| 15 | SOX6 | NA | 27010 | -0.021 | -0.3898 | No |
| 16 | SOX1 | NA | 30087 | -0.035 | -0.4385 | No |
| 17 | NKX2-2 | NA | 31365 | -0.041 | -0.4529 | No |
| 18 | SOX4 | NA | 32300 | -0.046 | -0.4600 | No |
| 19 | LHX3 | NA | 33117 | -0.050 | -0.4640 | No |
| 20 | LHX5 | NA | 33690 | -0.053 | -0.4629 | No |
| 21 | ISL2 | NA | 33709 | -0.053 | -0.4516 | No |
| 22 | GSX2 | NA | 34095 | -0.055 | -0.4466 | No |
| 23 | LMO4 | NA | 36017 | -0.066 | -0.4675 | No |
| 24 | MNX1 | NA | 37220 | -0.072 | -0.4738 | No |
| 25 | PHOX2A | NA | 38795 | -0.081 | -0.4850 | No |
| 26 | MDGA2 | NA | 38842 | -0.081 | -0.4682 | No |
| 27 | GDF7 | NA | 40569 | -0.091 | -0.4798 | No |
| 28 | SHH | NA | 43384 | -0.109 | -0.5075 | Yes |
| 29 | DMRT3 | NA | 44240 | -0.115 | -0.4980 | Yes |
| 30 | SCYL3 | NA | 45180 | -0.122 | -0.4885 | Yes |
| 31 | DLL4 | NA | 45773 | -0.127 | -0.4716 | Yes |
| 32 | DYNC2H1 | NA | 46122 | -0.130 | -0.4497 | Yes |
| 33 | ZC4H2 | NA | 46676 | -0.134 | -0.4305 | Yes |
| 34 | HOXC10 | NA | 46790 | -0.135 | -0.4030 | Yes |
| 35 | LBX1 | NA | 47342 | -0.140 | -0.3826 | Yes |
| 36 | DRAXIN | NA | 47717 | -0.143 | -0.3581 | Yes |
| 37 | LHX4 | NA | 48205 | -0.148 | -0.3348 | Yes |
| 38 | TCTN1 | NA | 49004 | -0.155 | -0.3156 | Yes |
| 39 | GLI2 | NA | 49153 | -0.156 | -0.2842 | Yes |
| 40 | MDGA1 | NA | 49245 | -0.157 | -0.2515 | Yes |
| 41 | WNT1 | NA | 49337 | -0.158 | -0.2187 | Yes |
| 42 | TAL1 | NA | 49754 | -0.162 | -0.1908 | Yes |
| 43 | HOXD10 | NA | 49875 | -0.164 | -0.1573 | Yes |
| 44 | IFT172 | NA | 50014 | -0.165 | -0.1237 | Yes |
| 45 | SUFU | NA | 50137 | -0.167 | -0.0896 | Yes |
| 46 | SOX13 | NA | 52084 | -0.193 | -0.0832 | Yes |
| 47 | CLN8 | NA | 52136 | -0.194 | -0.0418 | Yes |
| 48 | GATA2 | NA | 52360 | -0.197 | -0.0027 | Yes |
| 49 | PTCH1 | NA | 52477 | -0.200 | 0.0388 | Yes |