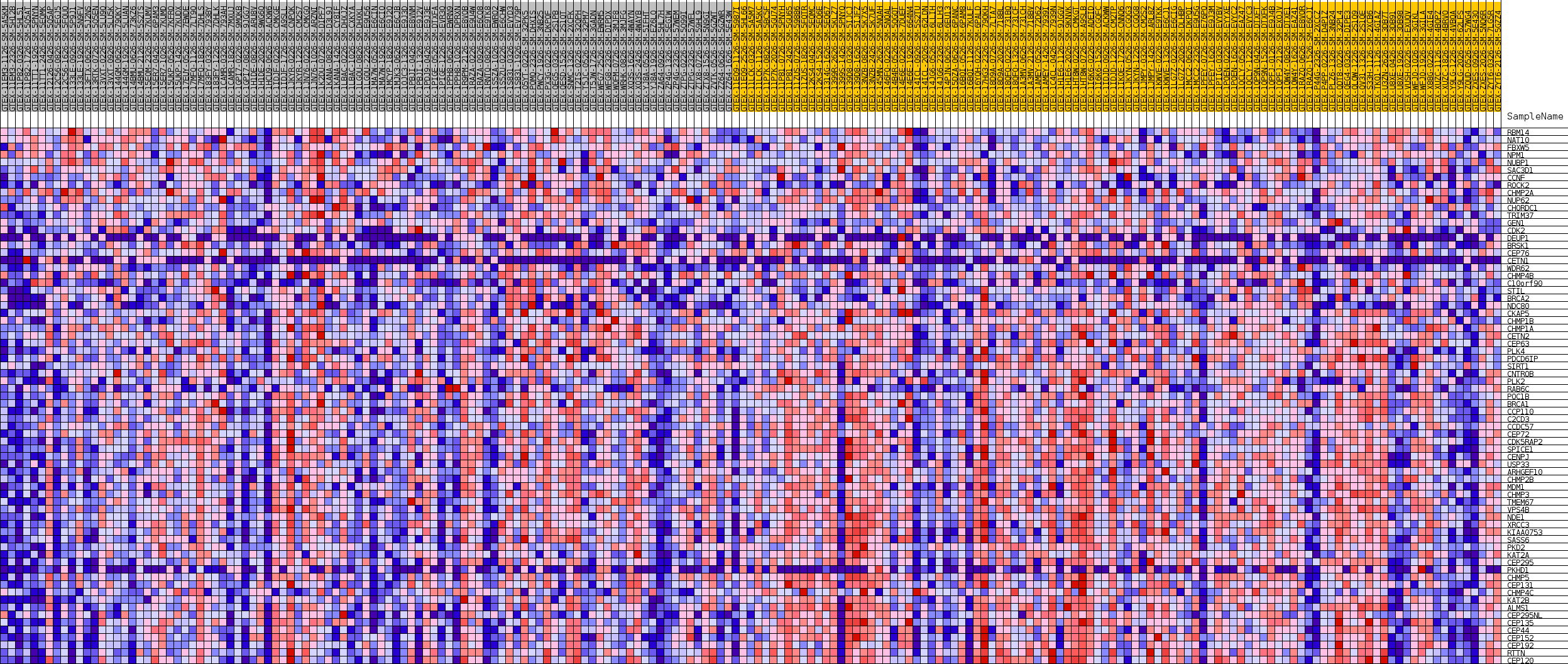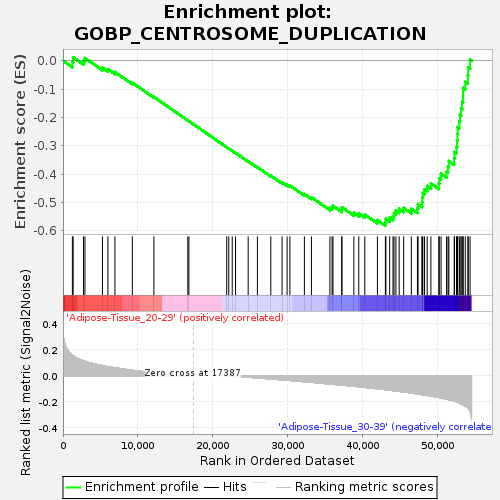
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_30-39 |
| GeneSet | GOBP_CENTROSOME_DUPLICATION |
| Enrichment Score (ES) | -0.5818519 |
| Normalized Enrichment Score (NES) | -1.5232986 |
| Nominal p-value | 0.05313093 |
| FDR q-value | 0.86144316 |
| FWER p-Value | 0.948 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | RBM14 | NA | 1252 | 0.158 | -0.0045 | No |
| 2 | NAT10 | NA | 1356 | 0.154 | 0.0115 | No |
| 3 | FBXW5 | NA | 2734 | 0.115 | -0.0004 | No |
| 4 | NPM1 | NA | 2935 | 0.111 | 0.0089 | No |
| 5 | NUBP1 | NA | 5267 | 0.078 | -0.0248 | No |
| 6 | SAC3D1 | NA | 6000 | 0.070 | -0.0301 | No |
| 7 | CCNF | NA | 6937 | 0.061 | -0.0401 | No |
| 8 | ROCK2 | NA | 9264 | 0.042 | -0.0779 | No |
| 9 | CHMP2A | NA | 12144 | 0.023 | -0.1280 | No |
| 10 | NUP62 | NA | 16679 | 0.003 | -0.2108 | No |
| 11 | CHORDC1 | NA | 16824 | 0.002 | -0.2131 | No |
| 12 | TRIM37 | NA | 21875 | -0.002 | -0.3055 | No |
| 13 | GEN1 | NA | 22149 | -0.003 | -0.3102 | No |
| 14 | CDK2 | NA | 22611 | -0.004 | -0.3181 | No |
| 15 | DEUP1 | NA | 23057 | -0.006 | -0.3256 | No |
| 16 | BRSK1 | NA | 24742 | -0.012 | -0.3552 | No |
| 17 | CEP76 | NA | 25974 | -0.017 | -0.3758 | No |
| 18 | CETN1 | NA | 27767 | -0.025 | -0.4058 | No |
| 19 | WDR62 | NA | 29268 | -0.032 | -0.4296 | No |
| 20 | CHMP4B | NA | 29941 | -0.035 | -0.4379 | No |
| 21 | C10orf90 | NA | 30315 | -0.036 | -0.4405 | No |
| 22 | STIL | NA | 32251 | -0.046 | -0.4706 | No |
| 23 | BRCA2 | NA | 33192 | -0.050 | -0.4820 | No |
| 24 | NDC80 | NA | 35663 | -0.064 | -0.5199 | No |
| 25 | CKAP5 | NA | 35951 | -0.065 | -0.5176 | No |
| 26 | CHMP1B | NA | 36075 | -0.066 | -0.5121 | No |
| 27 | CHMP1A | NA | 37245 | -0.072 | -0.5252 | No |
| 28 | CETN2 | NA | 37270 | -0.072 | -0.5172 | No |
| 29 | CEP63 | NA | 38862 | -0.081 | -0.5369 | No |
| 30 | PLK4 | NA | 39519 | -0.085 | -0.5390 | No |
| 31 | PDCD6IP | NA | 40324 | -0.090 | -0.5432 | No |
| 32 | SIRT1 | NA | 42010 | -0.100 | -0.5624 | No |
| 33 | CNTROB | NA | 43071 | -0.107 | -0.5693 | Yes |
| 34 | PLK2 | NA | 43148 | -0.108 | -0.5581 | Yes |
| 35 | RAB6C | NA | 43649 | -0.111 | -0.5543 | Yes |
| 36 | POC1B | NA | 44093 | -0.114 | -0.5491 | Yes |
| 37 | BRCA1 | NA | 44227 | -0.115 | -0.5381 | Yes |
| 38 | CCP110 | NA | 44484 | -0.117 | -0.5291 | Yes |
| 39 | C2CD3 | NA | 44918 | -0.121 | -0.5230 | Yes |
| 40 | CCDC57 | NA | 45519 | -0.125 | -0.5194 | Yes |
| 41 | CEP72 | NA | 46541 | -0.133 | -0.5226 | Yes |
| 42 | CDK5RAP2 | NA | 47358 | -0.140 | -0.5212 | Yes |
| 43 | SPICE1 | NA | 47442 | -0.141 | -0.5063 | Yes |
| 44 | CENPJ | NA | 47970 | -0.146 | -0.4990 | Yes |
| 45 | USP33 | NA | 47993 | -0.146 | -0.4824 | Yes |
| 46 | ARHGEF10 | NA | 48080 | -0.147 | -0.4669 | Yes |
| 47 | CHMP2B | NA | 48306 | -0.149 | -0.4537 | Yes |
| 48 | MDM1 | NA | 48658 | -0.152 | -0.4424 | Yes |
| 49 | CHMP3 | NA | 49157 | -0.156 | -0.4333 | Yes |
| 50 | TMEM67 | NA | 50206 | -0.167 | -0.4330 | Yes |
| 51 | VPS4B | NA | 50276 | -0.168 | -0.4147 | Yes |
| 52 | NDE1 | NA | 50512 | -0.171 | -0.3991 | Yes |
| 53 | XRCC3 | NA | 51249 | -0.180 | -0.3916 | Yes |
| 54 | KIAA0753 | NA | 51444 | -0.183 | -0.3738 | Yes |
| 55 | SASS6 | NA | 51556 | -0.185 | -0.3542 | Yes |
| 56 | PKD2 | NA | 52269 | -0.196 | -0.3445 | Yes |
| 57 | KAT2A | NA | 52308 | -0.196 | -0.3223 | Yes |
| 58 | CEP295 | NA | 52576 | -0.202 | -0.3036 | Yes |
| 59 | PKHD1 | NA | 52652 | -0.203 | -0.2813 | Yes |
| 60 | CHMP5 | NA | 52716 | -0.204 | -0.2586 | Yes |
| 61 | CEP131 | NA | 52727 | -0.205 | -0.2350 | Yes |
| 62 | CHMP4C | NA | 52948 | -0.210 | -0.2145 | Yes |
| 63 | KAT2B | NA | 53043 | -0.212 | -0.1915 | Yes |
| 64 | ALMS1 | NA | 53190 | -0.215 | -0.1691 | Yes |
| 65 | CEP295NL | NA | 53323 | -0.219 | -0.1460 | Yes |
| 66 | CEP135 | NA | 53450 | -0.222 | -0.1224 | Yes |
| 67 | CEP44 | NA | 53453 | -0.222 | -0.0964 | Yes |
| 68 | CEP152 | NA | 53751 | -0.233 | -0.0747 | Yes |
| 69 | CEP192 | NA | 54097 | -0.248 | -0.0521 | Yes |
| 70 | RTTN | NA | 54146 | -0.251 | -0.0237 | Yes |
| 71 | CEP120 | NA | 54393 | -0.273 | 0.0036 | Yes |