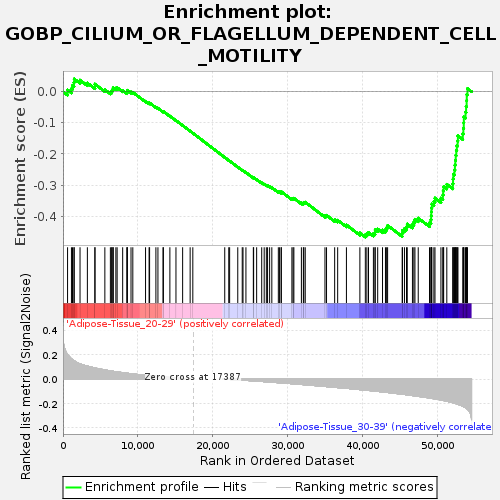
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_30-39 |
| GeneSet | GOBP_CILIUM_OR_FLAGELLUM_DEPENDENT_CELL_MOTILITY |
| Enrichment Score (ES) | -0.4671852 |
| Normalized Enrichment Score (NES) | -1.5695446 |
| Nominal p-value | 0.008 |
| FDR q-value | 0.9483667 |
| FWER p-Value | 0.926 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ADCY3 | NA | 610 | 0.199 | 0.0054 | No |
| 2 | APOB | NA | 1127 | 0.164 | 0.0097 | No |
| 3 | DNAAF2 | NA | 1269 | 0.157 | 0.0202 | No |
| 4 | CCNYL1 | NA | 1463 | 0.150 | 0.0292 | No |
| 5 | CFAP157 | NA | 1497 | 0.149 | 0.0410 | No |
| 6 | RNASE10 | NA | 2276 | 0.124 | 0.0371 | No |
| 7 | DNAJA1 | NA | 3256 | 0.105 | 0.0279 | No |
| 8 | PLTP | NA | 4265 | 0.090 | 0.0170 | No |
| 9 | EPPIN | NA | 4267 | 0.090 | 0.0245 | No |
| 10 | DRC1 | NA | 5592 | 0.074 | 0.0064 | No |
| 11 | GAS2L2 | NA | 6330 | 0.066 | -0.0016 | No |
| 12 | INTS13 | NA | 6504 | 0.065 | 0.0006 | No |
| 13 | PGAM4 | NA | 6551 | 0.064 | 0.0052 | No |
| 14 | PLA2G3 | NA | 6686 | 0.063 | 0.0080 | No |
| 15 | SLC9C1 | NA | 6709 | 0.063 | 0.0128 | No |
| 16 | RSPH9 | NA | 7065 | 0.060 | 0.0113 | No |
| 17 | NPHP4 | NA | 7231 | 0.058 | 0.0132 | No |
| 18 | DNAI1 | NA | 7972 | 0.052 | 0.0040 | No |
| 19 | AKAP4 | NA | 8548 | 0.047 | -0.0026 | No |
| 20 | RSPH4A | NA | 8578 | 0.047 | 0.0008 | No |
| 21 | DNAH17 | NA | 8584 | 0.047 | 0.0046 | No |
| 22 | SORD | NA | 9087 | 0.043 | -0.0010 | No |
| 23 | CFAP46 | NA | 9328 | 0.042 | -0.0019 | No |
| 24 | CFAP69 | NA | 11029 | 0.030 | -0.0306 | No |
| 25 | PRDM14 | NA | 11503 | 0.027 | -0.0370 | No |
| 26 | SMCP | NA | 11570 | 0.027 | -0.0360 | No |
| 27 | DRC7 | NA | 12411 | 0.022 | -0.0496 | No |
| 28 | CFAP20 | NA | 12685 | 0.020 | -0.0529 | No |
| 29 | DNAH11 | NA | 13364 | 0.017 | -0.0639 | No |
| 30 | DPCD | NA | 13413 | 0.017 | -0.0634 | No |
| 31 | GAPDHS | NA | 14281 | 0.013 | -0.0782 | No |
| 32 | MAFIP | NA | 15093 | 0.009 | -0.0924 | No |
| 33 | LDHC | NA | 15985 | 0.005 | -0.1083 | No |
| 34 | DNAH2 | NA | 16987 | 0.002 | -0.1265 | No |
| 35 | QRICH2 | NA | 17345 | 0.000 | -0.1330 | No |
| 36 | SLC26A8 | NA | 21611 | -0.001 | -0.2112 | No |
| 37 | DEFB1 | NA | 22147 | -0.003 | -0.2208 | No |
| 38 | ATP1A4 | NA | 22265 | -0.003 | -0.2227 | No |
| 39 | TPGS1 | NA | 23351 | -0.007 | -0.2421 | No |
| 40 | TEKT1 | NA | 23947 | -0.009 | -0.2523 | No |
| 41 | PRSS55 | NA | 24052 | -0.009 | -0.2534 | No |
| 42 | CCDC65 | NA | 24442 | -0.011 | -0.2597 | No |
| 43 | CATSPER3 | NA | 25432 | -0.014 | -0.2766 | No |
| 44 | CATSPERD | NA | 25471 | -0.015 | -0.2761 | No |
| 45 | ATP2B4 | NA | 25874 | -0.016 | -0.2821 | No |
| 46 | NME8 | NA | 26550 | -0.019 | -0.2929 | No |
| 47 | TNP1 | NA | 26882 | -0.021 | -0.2972 | No |
| 48 | DDX4 | NA | 27168 | -0.022 | -0.3006 | No |
| 49 | TEX101 | NA | 27333 | -0.023 | -0.3018 | No |
| 50 | TEKT2 | NA | 27608 | -0.024 | -0.3048 | No |
| 51 | TPPP2 | NA | 27902 | -0.025 | -0.3081 | No |
| 52 | SPEM1 | NA | 28771 | -0.029 | -0.3216 | No |
| 53 | IQCF1 | NA | 28936 | -0.030 | -0.3221 | No |
| 54 | TEKT5 | NA | 29156 | -0.031 | -0.3235 | No |
| 55 | TEKT4 | NA | 29164 | -0.031 | -0.3210 | No |
| 56 | MEIG1 | NA | 30584 | -0.038 | -0.3439 | No |
| 57 | CATSPER4 | NA | 30738 | -0.038 | -0.3435 | No |
| 58 | SLC9B2 | NA | 30848 | -0.039 | -0.3423 | No |
| 59 | ENKUR | NA | 31852 | -0.044 | -0.3571 | No |
| 60 | DNAH5 | NA | 32115 | -0.045 | -0.3581 | No |
| 61 | MKKS | NA | 32176 | -0.045 | -0.3554 | No |
| 62 | EFCAB9 | NA | 32366 | -0.046 | -0.3550 | No |
| 63 | ROPN1 | NA | 35002 | -0.060 | -0.3983 | No |
| 64 | NEURL1 | NA | 35210 | -0.061 | -0.3970 | No |
| 65 | CFAP43 | NA | 36298 | -0.067 | -0.4113 | No |
| 66 | RSPH6A | NA | 36700 | -0.069 | -0.4129 | No |
| 67 | CFAP65 | NA | 37869 | -0.075 | -0.4280 | No |
| 68 | CCDC40 | NA | 39667 | -0.086 | -0.4538 | No |
| 69 | ROPN1L | NA | 40395 | -0.090 | -0.4596 | Yes |
| 70 | TACR3 | NA | 40599 | -0.092 | -0.4557 | Yes |
| 71 | SEMG2 | NA | 40819 | -0.093 | -0.4520 | Yes |
| 72 | DNAH8 | NA | 41474 | -0.097 | -0.4559 | Yes |
| 73 | SEMG1 | NA | 41683 | -0.098 | -0.4514 | Yes |
| 74 | ARMC2 | NA | 41712 | -0.099 | -0.4437 | Yes |
| 75 | ASH1L | NA | 42047 | -0.101 | -0.4414 | Yes |
| 76 | ROPN1B | NA | 42692 | -0.105 | -0.4445 | Yes |
| 77 | DNAH3 | NA | 43085 | -0.107 | -0.4427 | Yes |
| 78 | GAS8 | NA | 43244 | -0.109 | -0.4365 | Yes |
| 79 | TSSK4 | NA | 43372 | -0.109 | -0.4297 | Yes |
| 80 | DNAH14 | NA | 45322 | -0.124 | -0.4552 | Yes |
| 81 | CCR6 | NA | 45357 | -0.124 | -0.4455 | Yes |
| 82 | TMF1 | NA | 45623 | -0.126 | -0.4398 | Yes |
| 83 | DNAH6 | NA | 45897 | -0.128 | -0.4341 | Yes |
| 84 | RGN | NA | 45995 | -0.129 | -0.4252 | Yes |
| 85 | SPEF2 | NA | 46690 | -0.134 | -0.4267 | Yes |
| 86 | CATSPER2 | NA | 46876 | -0.136 | -0.4187 | Yes |
| 87 | CFAP54 | NA | 47023 | -0.137 | -0.4099 | Yes |
| 88 | LRRC6 | NA | 47464 | -0.141 | -0.4062 | Yes |
| 89 | TAC4 | NA | 48971 | -0.155 | -0.4209 | Yes |
| 90 | TACR2 | NA | 49159 | -0.156 | -0.4113 | Yes |
| 91 | UBE2B | NA | 49192 | -0.157 | -0.3988 | Yes |
| 92 | CATSPER1 | NA | 49215 | -0.157 | -0.3861 | Yes |
| 93 | TTLL1 | NA | 49234 | -0.157 | -0.3733 | Yes |
| 94 | TACR1 | NA | 49287 | -0.158 | -0.3610 | Yes |
| 95 | TCTE1 | NA | 49556 | -0.160 | -0.3526 | Yes |
| 96 | BBS4 | NA | 49701 | -0.162 | -0.3417 | Yes |
| 97 | PRM3 | NA | 50496 | -0.171 | -0.3420 | Yes |
| 98 | EFHC2 | NA | 50753 | -0.174 | -0.3321 | Yes |
| 99 | TTC12 | NA | 50796 | -0.174 | -0.3183 | Yes |
| 100 | SPAG16 | NA | 50861 | -0.175 | -0.3049 | Yes |
| 101 | CACNA1I | NA | 51282 | -0.181 | -0.2975 | Yes |
| 102 | DNAH1 | NA | 52085 | -0.193 | -0.2961 | Yes |
| 103 | CFAP44 | NA | 52116 | -0.194 | -0.2804 | Yes |
| 104 | VPS13A | NA | 52152 | -0.194 | -0.2649 | Yes |
| 105 | IQCG | NA | 52306 | -0.196 | -0.2513 | Yes |
| 106 | TTLL9 | NA | 52366 | -0.198 | -0.2358 | Yes |
| 107 | BBS2 | NA | 52424 | -0.199 | -0.2202 | Yes |
| 108 | CCDC39 | NA | 52472 | -0.200 | -0.2044 | Yes |
| 109 | ING2 | NA | 52545 | -0.201 | -0.1889 | Yes |
| 110 | EFHC1 | NA | 52617 | -0.203 | -0.1733 | Yes |
| 111 | CEP131 | NA | 52727 | -0.205 | -0.1582 | Yes |
| 112 | DNAH7 | NA | 52764 | -0.205 | -0.1417 | Yes |
| 113 | FSIP2 | NA | 53418 | -0.221 | -0.1352 | Yes |
| 114 | CFAP206 | NA | 53495 | -0.224 | -0.1178 | Yes |
| 115 | TEKT3 | NA | 53547 | -0.226 | -0.0999 | Yes |
| 116 | CCSAP | NA | 53559 | -0.226 | -0.0812 | Yes |
| 117 | RFX3 | NA | 53784 | -0.234 | -0.0658 | Yes |
| 118 | INPP5B | NA | 53862 | -0.237 | -0.0474 | Yes |
| 119 | TTC21A | NA | 53929 | -0.240 | -0.0285 | Yes |
| 120 | SLC9B1 | NA | 53948 | -0.241 | -0.0088 | Yes |
| 121 | SLC22A16 | NA | 54043 | -0.246 | 0.0101 | Yes |