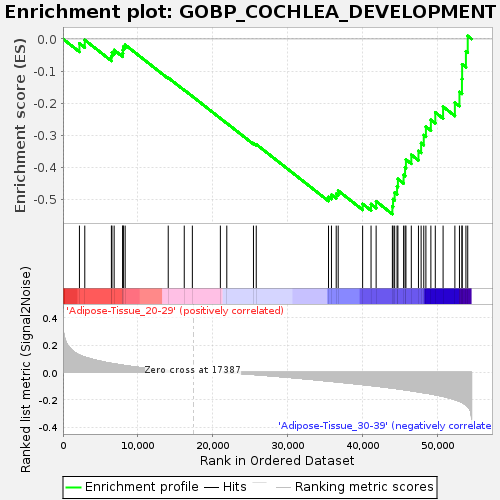
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_30-39 |
| GeneSet | GOBP_COCHLEA_DEVELOPMENT |
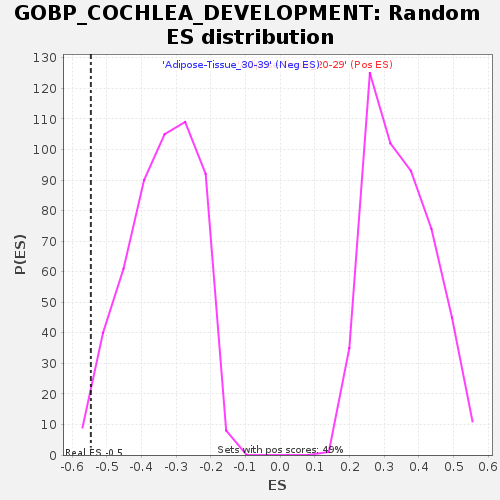
| Enrichment Score (ES) | -0.54593354 |
| Normalized Enrichment Score (NES) | -1.6030055 |
| Nominal p-value | 0.013618677 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.888 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | GABRA5 | NA | 2196 | 0.126 | -0.0137 | No |
| 2 | SOX9 | NA | 2910 | 0.111 | -0.0033 | No |
| 3 | GATA3 | NA | 6465 | 0.065 | -0.0547 | No |
| 4 | PTK7 | NA | 6540 | 0.065 | -0.0424 | No |
| 5 | GRHL3 | NA | 6834 | 0.062 | -0.0347 | No |
| 6 | POU3F4 | NA | 7975 | 0.052 | -0.0446 | No |
| 7 | CECR2 | NA | 8008 | 0.052 | -0.0342 | No |
| 8 | KCNK3 | NA | 8057 | 0.051 | -0.0242 | No |
| 9 | OPA1 | NA | 8304 | 0.049 | -0.0183 | No |
| 10 | SLC17A8 | NA | 14060 | 0.014 | -0.1209 | No |
| 11 | SIX1 | NA | 16201 | 0.005 | -0.1592 | No |
| 12 | PAX2 | NA | 17284 | 0.001 | -0.1789 | No |
| 13 | DCANP1 | NA | 21026 | 0.000 | -0.2475 | No |
| 14 | RPGRIP1L | NA | 21885 | -0.002 | -0.2628 | No |
| 15 | PVALB | NA | 25449 | -0.014 | -0.3250 | No |
| 16 | MCM2 | NA | 25820 | -0.016 | -0.3284 | No |
| 17 | DVL1 | NA | 35476 | -0.063 | -0.4922 | No |
| 18 | WNT5A | NA | 35870 | -0.065 | -0.4857 | No |
| 19 | DCHS1 | NA | 36505 | -0.068 | -0.4829 | No |
| 20 | CCNA2 | NA | 36770 | -0.070 | -0.4730 | No |
| 21 | MYO3A | NA | 40034 | -0.088 | -0.5142 | No |
| 22 | NEUROG1 | NA | 41159 | -0.095 | -0.5147 | No |
| 23 | CALB1 | NA | 41839 | -0.099 | -0.5062 | No |
| 24 | IFT20 | NA | 44009 | -0.114 | -0.5219 | Yes |
| 25 | DVL2 | NA | 44099 | -0.115 | -0.4993 | Yes |
| 26 | SOBP | NA | 44313 | -0.116 | -0.4787 | Yes |
| 27 | KCNK2 | NA | 44635 | -0.118 | -0.4595 | Yes |
| 28 | GABRB2 | NA | 44748 | -0.119 | -0.4364 | Yes |
| 29 | FZD2 | NA | 45508 | -0.125 | -0.4239 | Yes |
| 30 | MYO3B | NA | 45704 | -0.126 | -0.4007 | Yes |
| 31 | HOXA1 | NA | 45821 | -0.127 | -0.3760 | Yes |
| 32 | TBX18 | NA | 46539 | -0.133 | -0.3610 | Yes |
| 33 | HPN | NA | 47494 | -0.141 | -0.3486 | Yes |
| 34 | TBX1 | NA | 47850 | -0.145 | -0.3246 | Yes |
| 35 | CTHRC1 | NA | 48204 | -0.148 | -0.2998 | Yes |
| 36 | HEY2 | NA | 48488 | -0.150 | -0.2733 | Yes |
| 37 | GLI2 | NA | 49153 | -0.156 | -0.2524 | Yes |
| 38 | IFT27 | NA | 49744 | -0.162 | -0.2289 | Yes |
| 39 | HES1 | NA | 50785 | -0.174 | -0.2112 | Yes |
| 40 | GATA2 | NA | 52360 | -0.197 | -0.1983 | Yes |
| 41 | SLITRK6 | NA | 52972 | -0.210 | -0.1651 | Yes |
| 42 | EPHA4 | NA | 53305 | -0.218 | -0.1250 | Yes |
| 43 | ADAM10 | NA | 53330 | -0.219 | -0.0791 | Yes |
| 44 | TIFAB | NA | 53836 | -0.236 | -0.0385 | Yes |
| 45 | FRZB | NA | 54078 | -0.248 | 0.0094 | Yes |