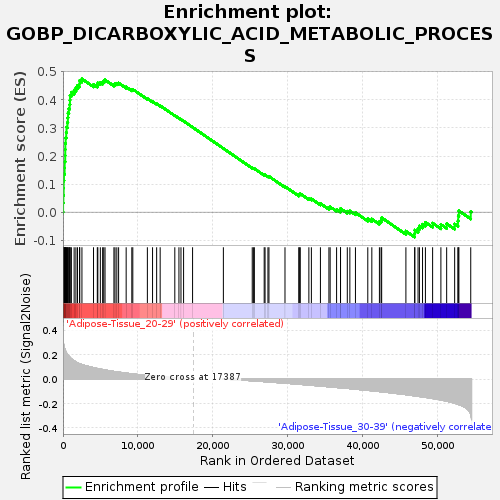
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_20-29 |
| GeneSet | GOBP_DICARBOXYLIC_ACID_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.47467303 |
| Normalized Enrichment Score (NES) | 1.4088086 |
| Nominal p-value | 0.108 |
| FDR q-value | 0.64827865 |
| FWER p-Value | 0.985 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ASS1 | NA | 11 | 0.358 | 0.0329 | Yes |
| 2 | FH | NA | 48 | 0.300 | 0.0600 | Yes |
| 3 | SDHB | NA | 84 | 0.284 | 0.0857 | Yes |
| 4 | SHMT1 | NA | 92 | 0.282 | 0.1117 | Yes |
| 5 | PCK1 | NA | 127 | 0.271 | 0.1361 | Yes |
| 6 | GOT1 | NA | 216 | 0.251 | 0.1577 | Yes |
| 7 | ALDH18A1 | NA | 224 | 0.249 | 0.1806 | Yes |
| 8 | GLUD2 | NA | 296 | 0.238 | 0.2013 | Yes |
| 9 | GLUL | NA | 302 | 0.237 | 0.2231 | Yes |
| 10 | MDH2 | NA | 313 | 0.235 | 0.2447 | Yes |
| 11 | FPGS | NA | 353 | 0.228 | 0.2651 | Yes |
| 12 | DLST | NA | 428 | 0.217 | 0.2838 | Yes |
| 13 | IDH2 | NA | 468 | 0.213 | 0.3028 | Yes |
| 14 | GPT2 | NA | 591 | 0.201 | 0.3191 | Yes |
| 15 | DLD | NA | 645 | 0.196 | 0.3363 | Yes |
| 16 | MTHFD1L | NA | 677 | 0.194 | 0.3536 | Yes |
| 17 | ATIC | NA | 782 | 0.186 | 0.3690 | Yes |
| 18 | SHMT2 | NA | 897 | 0.179 | 0.3834 | Yes |
| 19 | ALDH1L1 | NA | 927 | 0.177 | 0.3993 | Yes |
| 20 | OGDH | NA | 943 | 0.176 | 0.4153 | Yes |
| 21 | MTHFD1 | NA | 1119 | 0.164 | 0.4273 | Yes |
| 22 | ACLY | NA | 1487 | 0.149 | 0.4343 | Yes |
| 23 | ACOT4 | NA | 1725 | 0.140 | 0.4429 | Yes |
| 24 | SLC7A11 | NA | 1933 | 0.133 | 0.4514 | Yes |
| 25 | SDHA | NA | 2223 | 0.125 | 0.4577 | Yes |
| 26 | MRPS36 | NA | 2226 | 0.125 | 0.4692 | Yes |
| 27 | L2HGDH | NA | 2530 | 0.119 | 0.4747 | Yes |
| 28 | NIT2 | NA | 4078 | 0.093 | 0.4549 | No |
| 29 | GCLM | NA | 4584 | 0.086 | 0.4536 | No |
| 30 | SLC19A1 | NA | 4654 | 0.085 | 0.4602 | No |
| 31 | ME1 | NA | 4989 | 0.081 | 0.4616 | No |
| 32 | PRODH2 | NA | 5297 | 0.078 | 0.4632 | No |
| 33 | KYAT3 | NA | 5443 | 0.076 | 0.4675 | No |
| 34 | MDH1 | NA | 5615 | 0.074 | 0.4712 | No |
| 35 | MTRR | NA | 6804 | 0.062 | 0.4552 | No |
| 36 | SLC25A32 | NA | 6949 | 0.061 | 0.4582 | No |
| 37 | PCK2 | NA | 7193 | 0.059 | 0.4591 | No |
| 38 | MTHFD2 | NA | 7430 | 0.057 | 0.4601 | No |
| 39 | DHFR | NA | 8439 | 0.048 | 0.4460 | No |
| 40 | MTHFD2L | NA | 9194 | 0.043 | 0.4361 | No |
| 41 | IDH1 | NA | 9327 | 0.042 | 0.4376 | No |
| 42 | NAGS | NA | 11273 | 0.028 | 0.4045 | No |
| 43 | DHFRP1 | NA | 11965 | 0.025 | 0.3941 | No |
| 44 | GAD2 | NA | 12513 | 0.021 | 0.3860 | No |
| 45 | ME2 | NA | 12983 | 0.019 | 0.3792 | No |
| 46 | SDHAF3 | NA | 14939 | 0.010 | 0.3442 | No |
| 47 | ACSF3 | NA | 15490 | 0.007 | 0.3348 | No |
| 48 | GLUD1 | NA | 15763 | 0.006 | 0.3304 | No |
| 49 | ATCAY | NA | 16112 | 0.005 | 0.3245 | No |
| 50 | SUCLG2 | NA | 17313 | 0.000 | 0.3025 | No |
| 51 | HAL | NA | 21428 | -0.000 | 0.2270 | No |
| 52 | KYNU | NA | 25284 | -0.014 | 0.1576 | No |
| 53 | GOT1L1 | NA | 25438 | -0.014 | 0.1561 | No |
| 54 | PHYH | NA | 25569 | -0.015 | 0.1551 | No |
| 55 | OAT | NA | 26881 | -0.021 | 0.1329 | No |
| 56 | DDO | NA | 26997 | -0.021 | 0.1328 | No |
| 57 | GCLC | NA | 27384 | -0.023 | 0.1278 | No |
| 58 | MTHFS | NA | 27518 | -0.023 | 0.1276 | No |
| 59 | AMDHD1 | NA | 29660 | -0.034 | 0.0914 | No |
| 60 | GOT2 | NA | 31502 | -0.042 | 0.0615 | No |
| 61 | MDH1B | NA | 31580 | -0.042 | 0.0640 | No |
| 62 | TH | NA | 31704 | -0.043 | 0.0657 | No |
| 63 | ALDH5A1 | NA | 32851 | -0.049 | 0.0492 | No |
| 64 | ME3 | NA | 33187 | -0.050 | 0.0478 | No |
| 65 | MTHFR | NA | 34396 | -0.057 | 0.0309 | No |
| 66 | FOLR1 | NA | 35530 | -0.063 | 0.0159 | No |
| 67 | TAT | NA | 35694 | -0.064 | 0.0188 | No |
| 68 | QPRT | NA | 36556 | -0.068 | 0.0094 | No |
| 69 | ACOT8 | NA | 37080 | -0.071 | 0.0064 | No |
| 70 | UROC1 | NA | 37082 | -0.071 | 0.0130 | No |
| 71 | ALDH1L2 | NA | 37986 | -0.076 | 0.0034 | No |
| 72 | ACMSD | NA | 38329 | -0.078 | 0.0044 | No |
| 73 | FOLR2 | NA | 39075 | -0.083 | -0.0016 | No |
| 74 | STAR | NA | 40724 | -0.092 | -0.0233 | No |
| 75 | LIPF | NA | 41264 | -0.096 | -0.0244 | No |
| 76 | KMO | NA | 42277 | -0.102 | -0.0335 | No |
| 77 | NR1H4 | NA | 42498 | -0.103 | -0.0279 | No |
| 78 | ASPA | NA | 42579 | -0.104 | -0.0198 | No |
| 79 | D2HGDH | NA | 45816 | -0.127 | -0.0674 | No |
| 80 | GAD1 | NA | 46984 | -0.137 | -0.0761 | No |
| 81 | FTCD | NA | 47010 | -0.137 | -0.0639 | No |
| 82 | HOGA1 | NA | 47439 | -0.141 | -0.0587 | No |
| 83 | SLC46A1 | NA | 47624 | -0.142 | -0.0489 | No |
| 84 | ALDH4A1 | NA | 48039 | -0.146 | -0.0430 | No |
| 85 | GLS | NA | 48423 | -0.150 | -0.0361 | No |
| 86 | HAAO | NA | 49386 | -0.159 | -0.0391 | No |
| 87 | PRODH | NA | 50492 | -0.171 | -0.0436 | No |
| 88 | DHFR2 | NA | 51263 | -0.181 | -0.0410 | No |
| 89 | GGT1 | NA | 52331 | -0.197 | -0.0423 | No |
| 90 | AADAT | NA | 52770 | -0.205 | -0.0313 | No |
| 91 | ADHFE1 | NA | 52809 | -0.206 | -0.0129 | No |
| 92 | SLC25A12 | NA | 52897 | -0.208 | 0.0048 | No |
| 93 | GLS2 | NA | 54480 | -0.284 | 0.0020 | No |