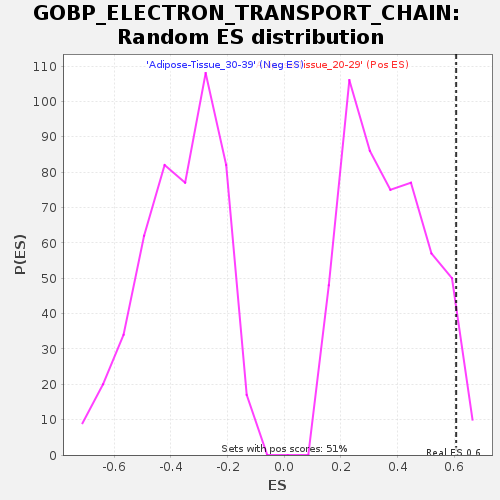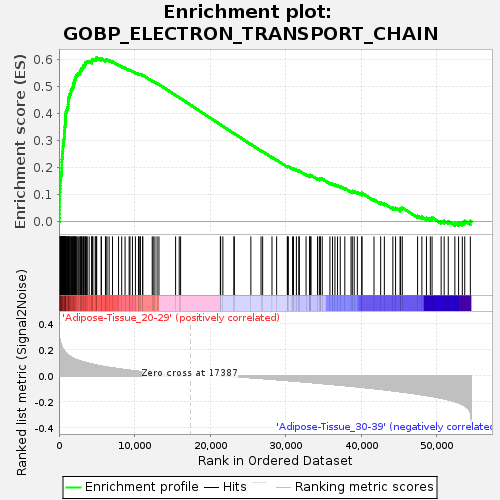
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_20-29 |
| GeneSet | GOBP_ELECTRON_TRANSPORT_CHAIN |
| Enrichment Score (ES) | 0.60677093 |
| Normalized Enrichment Score (NES) | 1.6626625 |
| Nominal p-value | 0.05108055 |
| FDR q-value | 0.225108 |
| FWER p-Value | 0.804 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | NDUFS2 | NA | 42 | 0.301 | 0.0136 | Yes |
| 2 | CYC1 | NA | 55 | 0.297 | 0.0277 | Yes |
| 3 | SLC25A22 | NA | 61 | 0.295 | 0.0417 | Yes |
| 4 | COX10 | NA | 69 | 0.291 | 0.0555 | Yes |
| 5 | NDUFAB1 | NA | 80 | 0.286 | 0.0690 | Yes |
| 6 | SDHB | NA | 84 | 0.284 | 0.0826 | Yes |
| 7 | NDUFAF2 | NA | 101 | 0.279 | 0.0956 | Yes |
| 8 | UQCRHL | NA | 110 | 0.276 | 0.1087 | Yes |
| 9 | UQCRFS1 | NA | 139 | 0.267 | 0.1209 | Yes |
| 10 | COX5A | NA | 160 | 0.260 | 0.1330 | Yes |
| 11 | CYB5A | NA | 182 | 0.256 | 0.1449 | Yes |
| 12 | UQCRH | NA | 191 | 0.255 | 0.1569 | Yes |
| 13 | UQCRFS1P1 | NA | 233 | 0.247 | 0.1680 | Yes |
| 14 | AIFM2 | NA | 311 | 0.235 | 0.1779 | Yes |
| 15 | COX8A | NA | 355 | 0.228 | 0.1880 | Yes |
| 16 | NDUFA8 | NA | 359 | 0.227 | 0.1988 | Yes |
| 17 | NDUFS5 | NA | 370 | 0.224 | 0.2093 | Yes |
| 18 | ETFA | NA | 371 | 0.224 | 0.2201 | Yes |
| 19 | NDUFA6 | NA | 385 | 0.223 | 0.2305 | Yes |
| 20 | CYP1A2 | NA | 426 | 0.217 | 0.2402 | Yes |
| 21 | CYCS | NA | 449 | 0.215 | 0.2501 | Yes |
| 22 | UQCRC2 | NA | 481 | 0.211 | 0.2596 | Yes |
| 23 | NDUFA9 | NA | 512 | 0.209 | 0.2690 | Yes |
| 24 | CIAPIN1 | NA | 517 | 0.208 | 0.2789 | Yes |
| 25 | SCO2 | NA | 584 | 0.201 | 0.2873 | Yes |
| 26 | COX7B | NA | 601 | 0.200 | 0.2966 | Yes |
| 27 | DLD | NA | 645 | 0.196 | 0.3052 | Yes |
| 28 | UQCRC1 | NA | 698 | 0.192 | 0.3135 | Yes |
| 29 | COX7A2 | NA | 706 | 0.192 | 0.3225 | Yes |
| 30 | COA6 | NA | 714 | 0.191 | 0.3315 | Yes |
| 31 | MAOB | NA | 745 | 0.189 | 0.3400 | Yes |
| 32 | NDUFS1 | NA | 761 | 0.188 | 0.3488 | Yes |
| 33 | COX7C | NA | 806 | 0.184 | 0.3568 | Yes |
| 34 | NDUFAF1 | NA | 832 | 0.183 | 0.3651 | Yes |
| 35 | NDUFS6 | NA | 842 | 0.182 | 0.3736 | Yes |
| 36 | NDUFC2 | NA | 864 | 0.180 | 0.3819 | Yes |
| 37 | MYBBP1A | NA | 874 | 0.180 | 0.3903 | Yes |
| 38 | ETFDH | NA | 888 | 0.179 | 0.3986 | Yes |
| 39 | COX5B | NA | 930 | 0.177 | 0.4063 | Yes |
| 40 | COX6A1 | NA | 1008 | 0.172 | 0.4131 | Yes |
| 41 | COX6B1 | NA | 1118 | 0.165 | 0.4190 | Yes |
| 42 | NDUFV2 | NA | 1184 | 0.162 | 0.4256 | Yes |
| 43 | NDUFB9 | NA | 1210 | 0.160 | 0.4328 | Yes |
| 44 | NDUFB6 | NA | 1229 | 0.159 | 0.4401 | Yes |
| 45 | NDUFA1 | NA | 1232 | 0.159 | 0.4477 | Yes |
| 46 | UQCRQ | NA | 1262 | 0.158 | 0.4547 | Yes |
| 47 | NDUFA4 | NA | 1317 | 0.156 | 0.4611 | Yes |
| 48 | GPD1 | NA | 1419 | 0.151 | 0.4665 | Yes |
| 49 | GLRX2 | NA | 1442 | 0.150 | 0.4733 | Yes |
| 50 | PARK7 | NA | 1571 | 0.146 | 0.4779 | Yes |
| 51 | NDUFA7 | NA | 1580 | 0.145 | 0.4847 | Yes |
| 52 | UQCR10 | NA | 1669 | 0.142 | 0.4899 | Yes |
| 53 | ETFB | NA | 1778 | 0.138 | 0.4945 | Yes |
| 54 | COQ9 | NA | 1858 | 0.135 | 0.4996 | Yes |
| 55 | DEGS1 | NA | 1878 | 0.135 | 0.5057 | Yes |
| 56 | CHCHD2 | NA | 1884 | 0.134 | 0.5120 | Yes |
| 57 | QDPR | NA | 2042 | 0.129 | 0.5153 | Yes |
| 58 | NDUFB2 | NA | 2084 | 0.128 | 0.5207 | Yes |
| 59 | MT-CO1 | NA | 2118 | 0.127 | 0.5262 | Yes |
| 60 | NDUFV3 | NA | 2156 | 0.126 | 0.5316 | Yes |
| 61 | SDHA | NA | 2223 | 0.125 | 0.5363 | Yes |
| 62 | NDUFB3 | NA | 2295 | 0.124 | 0.5410 | Yes |
| 63 | GHITM | NA | 2408 | 0.121 | 0.5447 | Yes |
| 64 | MT-ND6 | NA | 2559 | 0.118 | 0.5476 | Yes |
| 65 | NDUFS3 | NA | 2781 | 0.114 | 0.5490 | Yes |
| 66 | NQO2 | NA | 2791 | 0.114 | 0.5543 | Yes |
| 67 | NDUFB8 | NA | 2814 | 0.113 | 0.5593 | Yes |
| 68 | COX6C | NA | 2950 | 0.111 | 0.5621 | Yes |
| 69 | MT-ND5 | NA | 2990 | 0.110 | 0.5666 | Yes |
| 70 | NDUFB11 | NA | 3141 | 0.107 | 0.5690 | Yes |
| 71 | NDUFA13 | NA | 3239 | 0.106 | 0.5723 | Yes |
| 72 | NDUFA12 | NA | 3245 | 0.106 | 0.5773 | Yes |
| 73 | MT-ND4 | NA | 3412 | 0.103 | 0.5791 | Yes |
| 74 | COX4I1 | NA | 3492 | 0.102 | 0.5826 | Yes |
| 75 | DHRS3 | NA | 3493 | 0.102 | 0.5874 | Yes |
| 76 | MT-ND4L | NA | 3628 | 0.099 | 0.5897 | Yes |
| 77 | NDUFS8 | NA | 3741 | 0.098 | 0.5923 | Yes |
| 78 | LOXL2 | NA | 3989 | 0.094 | 0.5923 | Yes |
| 79 | UQCC3 | NA | 4344 | 0.089 | 0.5901 | Yes |
| 80 | AOC2 | NA | 4350 | 0.089 | 0.5942 | Yes |
| 81 | IDH3B | NA | 4378 | 0.089 | 0.5980 | Yes |
| 82 | NDUFS7 | NA | 4492 | 0.087 | 0.6001 | Yes |
| 83 | UQCR11 | NA | 4792 | 0.084 | 0.5986 | Yes |
| 84 | COX7B2 | NA | 4907 | 0.082 | 0.6004 | Yes |
| 85 | NDUFA10 | NA | 4915 | 0.082 | 0.6042 | Yes |
| 86 | ME1 | NA | 4989 | 0.081 | 0.6068 | Yes |
| 87 | BID | NA | 5583 | 0.074 | 0.5994 | No |
| 88 | SDHAF2 | NA | 5610 | 0.074 | 0.6025 | No |
| 89 | SDHC | NA | 6151 | 0.068 | 0.5958 | No |
| 90 | NDUFA3 | NA | 6211 | 0.068 | 0.5980 | No |
| 91 | NDUFB10 | NA | 6353 | 0.066 | 0.5986 | No |
| 92 | NDUFB7 | NA | 6634 | 0.064 | 0.5965 | No |
| 93 | PHGDH | NA | 7085 | 0.060 | 0.5910 | No |
| 94 | CYB5B | NA | 7884 | 0.053 | 0.5789 | No |
| 95 | NDUFB1 | NA | 8300 | 0.049 | 0.5737 | No |
| 96 | SDHD | NA | 8747 | 0.046 | 0.5677 | No |
| 97 | NDUFA2 | NA | 9287 | 0.042 | 0.5597 | No |
| 98 | CCNB1 | NA | 9393 | 0.041 | 0.5598 | No |
| 99 | COX7A2P2 | NA | 9721 | 0.039 | 0.5556 | No |
| 100 | GLRX | NA | 10114 | 0.036 | 0.5502 | No |
| 101 | GLDC | NA | 10520 | 0.033 | 0.5443 | No |
| 102 | NCF1 | NA | 10539 | 0.033 | 0.5456 | No |
| 103 | NDUFS4 | NA | 10698 | 0.032 | 0.5442 | No |
| 104 | P4HA2 | NA | 10777 | 0.032 | 0.5443 | No |
| 105 | MT-CO2 | NA | 11073 | 0.030 | 0.5403 | No |
| 106 | ALDH2 | NA | 11086 | 0.030 | 0.5415 | No |
| 107 | DNAJC15 | NA | 12342 | 0.022 | 0.5195 | No |
| 108 | MTCO2P12 | NA | 12469 | 0.022 | 0.5182 | No |
| 109 | DGUOK | NA | 12697 | 0.020 | 0.5150 | No |
| 110 | ME2 | NA | 12983 | 0.019 | 0.5107 | No |
| 111 | CYP19A1 | NA | 13231 | 0.018 | 0.5070 | No |
| 112 | WDR93 | NA | 15434 | 0.007 | 0.4669 | No |
| 113 | NDUFB4 | NA | 15916 | 0.006 | 0.4583 | No |
| 114 | NDUFB5 | NA | 16097 | 0.005 | 0.4553 | No |
| 115 | UQCRB | NA | 21357 | -0.000 | 0.3586 | No |
| 116 | GPD2 | NA | 21399 | -0.000 | 0.3579 | No |
| 117 | MT-CO3 | NA | 21694 | -0.002 | 0.3526 | No |
| 118 | HSD17B6 | NA | 23149 | -0.006 | 0.3261 | No |
| 119 | AKR1A1 | NA | 23209 | -0.006 | 0.3253 | No |
| 120 | NDUFV1 | NA | 25399 | -0.014 | 0.2858 | No |
| 121 | AKR7A2 | NA | 26746 | -0.020 | 0.2620 | No |
| 122 | NDUFC1 | NA | 26958 | -0.021 | 0.2591 | No |
| 123 | COX15 | NA | 28194 | -0.027 | 0.2377 | No |
| 124 | NDUFA11 | NA | 28809 | -0.029 | 0.2278 | No |
| 125 | AOX1 | NA | 30222 | -0.036 | 0.2036 | No |
| 126 | DHDH | NA | 30256 | -0.036 | 0.2047 | No |
| 127 | C15orf48 | NA | 30359 | -0.037 | 0.2046 | No |
| 128 | PINK1 | NA | 30921 | -0.039 | 0.1962 | No |
| 129 | CDK1 | NA | 31040 | -0.040 | 0.1959 | No |
| 130 | NDUFA5 | NA | 31427 | -0.042 | 0.1908 | No |
| 131 | ADH5 | NA | 31748 | -0.043 | 0.1870 | No |
| 132 | AKR1C4 | NA | 31813 | -0.044 | 0.1879 | No |
| 133 | GSR | NA | 32707 | -0.048 | 0.1738 | No |
| 134 | FDX1 | NA | 33172 | -0.050 | 0.1677 | No |
| 135 | COX8C | NA | 33202 | -0.050 | 0.1696 | No |
| 136 | MT-CYB | NA | 33330 | -0.051 | 0.1697 | No |
| 137 | MIR210 | NA | 33341 | -0.051 | 0.1719 | No |
| 138 | MT-ND1 | NA | 34219 | -0.056 | 0.1585 | No |
| 139 | COX7A1 | NA | 34460 | -0.057 | 0.1568 | No |
| 140 | ASPH | NA | 34569 | -0.058 | 0.1576 | No |
| 141 | FDX2 | NA | 34592 | -0.058 | 0.1600 | No |
| 142 | SLC25A18 | NA | 34848 | -0.059 | 0.1581 | No |
| 143 | NDUFA4L2 | NA | 35867 | -0.065 | 0.1425 | No |
| 144 | COX6A2 | NA | 36212 | -0.067 | 0.1394 | No |
| 145 | ETFRF1 | NA | 36529 | -0.068 | 0.1369 | No |
| 146 | CYBB | NA | 36872 | -0.070 | 0.1339 | No |
| 147 | SLC25A13 | NA | 37233 | -0.072 | 0.1308 | No |
| 148 | COX7A2L | NA | 37835 | -0.075 | 0.1233 | No |
| 149 | AKR7A3 | NA | 38659 | -0.080 | 0.1121 | No |
| 150 | SURF1 | NA | 38823 | -0.081 | 0.1129 | No |
| 151 | SNCA | NA | 39076 | -0.083 | 0.1123 | No |
| 152 | NCF2 | NA | 39497 | -0.085 | 0.1086 | No |
| 153 | COX4I2 | NA | 40080 | -0.089 | 0.1022 | No |
| 154 | IMMP2L | NA | 40105 | -0.089 | 0.1060 | No |
| 155 | COX11 | NA | 41699 | -0.099 | 0.0814 | No |
| 156 | DMGDH | NA | 42580 | -0.104 | 0.0702 | No |
| 157 | MT-ND3 | NA | 43070 | -0.107 | 0.0664 | No |
| 158 | STEAP4 | NA | 44196 | -0.115 | 0.0512 | No |
| 159 | CYBA | NA | 44556 | -0.118 | 0.0502 | No |
| 160 | PPARGC1A | NA | 45150 | -0.122 | 0.0452 | No |
| 161 | GPX2 | NA | 45221 | -0.123 | 0.0498 | No |
| 162 | AKR1B1 | NA | 45444 | -0.125 | 0.0517 | No |
| 163 | NOX4 | NA | 47469 | -0.141 | 0.0212 | No |
| 164 | ALDH4A1 | NA | 48039 | -0.146 | 0.0178 | No |
| 165 | IDO1 | NA | 48642 | -0.151 | 0.0140 | No |
| 166 | CYB561 | NA | 49156 | -0.156 | 0.0120 | No |
| 167 | HAAO | NA | 49386 | -0.159 | 0.0154 | No |
| 168 | SRD5A1 | NA | 50588 | -0.172 | 0.0016 | No |
| 169 | RDH16 | NA | 50988 | -0.177 | 0.0027 | No |
| 170 | ISCU | NA | 51531 | -0.184 | 0.0016 | No |
| 171 | POLG2 | NA | 52404 | -0.199 | -0.0049 | No |
| 172 | SLC25A12 | NA | 52897 | -0.208 | -0.0040 | No |
| 173 | XDH | NA | 53390 | -0.221 | -0.0025 | No |
| 174 | MT-ND2 | NA | 53712 | -0.232 | 0.0027 | No |
| 175 | TAZ | NA | 54454 | -0.280 | 0.0025 | No |