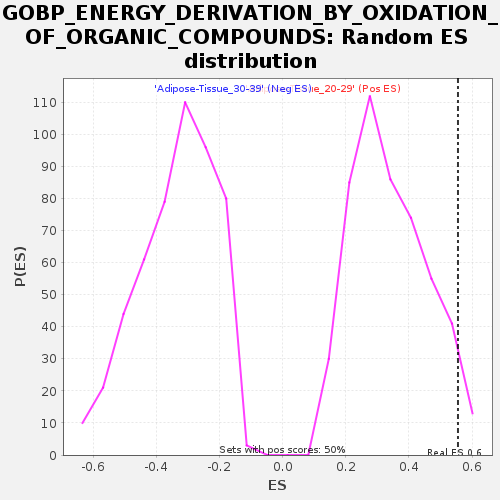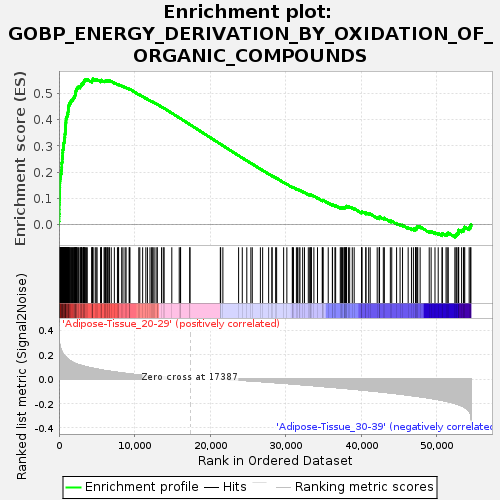
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_20-29 |
| GeneSet | GOBP_ENERGY_DERIVATION_BY_OXIDATION_OF_ORGANIC_COMPOUNDS |
| Enrichment Score (ES) | 0.55528766 |
| Normalized Enrichment Score (NES) | 1.6293503 |
| Nominal p-value | 0.048387095 |
| FDR q-value | 0.2622213 |
| FWER p-Value | 0.852 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | IGF1 | NA | 1 | 0.430 | 0.0126 | Yes |
| 2 | GFPT1 | NA | 23 | 0.324 | 0.0217 | Yes |
| 3 | PGM1 | NA | 25 | 0.322 | 0.0312 | Yes |
| 4 | CS | NA | 30 | 0.317 | 0.0404 | Yes |
| 5 | NDUFS2 | NA | 42 | 0.301 | 0.0490 | Yes |
| 6 | PDHA1 | NA | 43 | 0.301 | 0.0579 | Yes |
| 7 | FH | NA | 48 | 0.300 | 0.0666 | Yes |
| 8 | AKT1 | NA | 49 | 0.299 | 0.0753 | Yes |
| 9 | CYC1 | NA | 55 | 0.297 | 0.0840 | Yes |
| 10 | SLC25A22 | NA | 61 | 0.295 | 0.0925 | Yes |
| 11 | COX10 | NA | 69 | 0.291 | 0.1010 | Yes |
| 12 | NDUFAB1 | NA | 80 | 0.286 | 0.1092 | Yes |
| 13 | SDHB | NA | 84 | 0.284 | 0.1174 | Yes |
| 14 | GYS1 | NA | 89 | 0.283 | 0.1257 | Yes |
| 15 | DLAT | NA | 94 | 0.281 | 0.1338 | Yes |
| 16 | PRELID1 | NA | 99 | 0.279 | 0.1420 | Yes |
| 17 | UQCRHL | NA | 110 | 0.276 | 0.1499 | Yes |
| 18 | ACO1 | NA | 114 | 0.275 | 0.1579 | Yes |
| 19 | IDH3A | NA | 124 | 0.272 | 0.1657 | Yes |
| 20 | UQCRFS1 | NA | 139 | 0.267 | 0.1733 | Yes |
| 21 | COX5A | NA | 160 | 0.260 | 0.1805 | Yes |
| 22 | UQCRH | NA | 191 | 0.255 | 0.1874 | Yes |
| 23 | UQCRFS1P1 | NA | 233 | 0.247 | 0.1939 | Yes |
| 24 | AIFM2 | NA | 311 | 0.235 | 0.1994 | Yes |
| 25 | MDH2 | NA | 313 | 0.235 | 0.2063 | Yes |
| 26 | COX8A | NA | 355 | 0.228 | 0.2122 | Yes |
| 27 | NDUFA8 | NA | 359 | 0.227 | 0.2188 | Yes |
| 28 | NDUFS5 | NA | 370 | 0.224 | 0.2252 | Yes |
| 29 | ETFA | NA | 371 | 0.224 | 0.2318 | Yes |
| 30 | NDUFA6 | NA | 385 | 0.223 | 0.2381 | Yes |
| 31 | CYP1A2 | NA | 426 | 0.217 | 0.2437 | Yes |
| 32 | DLST | NA | 428 | 0.217 | 0.2501 | Yes |
| 33 | CYCS | NA | 449 | 0.215 | 0.2560 | Yes |
| 34 | IDH2 | NA | 468 | 0.213 | 0.2619 | Yes |
| 35 | UQCRC2 | NA | 481 | 0.211 | 0.2679 | Yes |
| 36 | ACO2 | NA | 483 | 0.211 | 0.2741 | Yes |
| 37 | ADRB3 | NA | 509 | 0.209 | 0.2797 | Yes |
| 38 | NDUFA9 | NA | 512 | 0.209 | 0.2858 | Yes |
| 39 | SCO2 | NA | 584 | 0.201 | 0.2904 | Yes |
| 40 | COX7B | NA | 601 | 0.200 | 0.2960 | Yes |
| 41 | IRS1 | NA | 605 | 0.199 | 0.3018 | Yes |
| 42 | STBD1 | NA | 612 | 0.199 | 0.3075 | Yes |
| 43 | DLD | NA | 645 | 0.196 | 0.3127 | Yes |
| 44 | UQCRC1 | NA | 698 | 0.192 | 0.3173 | Yes |
| 45 | COA6 | NA | 714 | 0.191 | 0.3227 | Yes |
| 46 | STK40 | NA | 736 | 0.190 | 0.3278 | Yes |
| 47 | NDUFS1 | NA | 761 | 0.188 | 0.3329 | Yes |
| 48 | GBE1 | NA | 779 | 0.186 | 0.3381 | Yes |
| 49 | PDHB | NA | 787 | 0.186 | 0.3434 | Yes |
| 50 | COX7C | NA | 806 | 0.184 | 0.3485 | Yes |
| 51 | PYGM | NA | 821 | 0.184 | 0.3536 | Yes |
| 52 | NDUFAF1 | NA | 832 | 0.183 | 0.3588 | Yes |
| 53 | NDUFS6 | NA | 842 | 0.182 | 0.3640 | Yes |
| 54 | NDUFC2 | NA | 864 | 0.180 | 0.3689 | Yes |
| 55 | MYBBP1A | NA | 874 | 0.180 | 0.3740 | Yes |
| 56 | ETFDH | NA | 888 | 0.179 | 0.3790 | Yes |
| 57 | SHMT2 | NA | 897 | 0.179 | 0.3841 | Yes |
| 58 | TRAP1 | NA | 911 | 0.178 | 0.3891 | Yes |
| 59 | PNPT1 | NA | 923 | 0.177 | 0.3940 | Yes |
| 60 | COX5B | NA | 930 | 0.177 | 0.3991 | Yes |
| 61 | OGDH | NA | 943 | 0.176 | 0.4041 | Yes |
| 62 | COX6A1 | NA | 1008 | 0.172 | 0.4079 | Yes |
| 63 | SELENOS | NA | 1046 | 0.169 | 0.4122 | Yes |
| 64 | VCP | NA | 1092 | 0.166 | 0.4162 | Yes |
| 65 | COX6B1 | NA | 1118 | 0.165 | 0.4206 | Yes |
| 66 | NDUFV2 | NA | 1184 | 0.162 | 0.4241 | Yes |
| 67 | PHLDA2 | NA | 1188 | 0.161 | 0.4288 | Yes |
| 68 | NDUFB9 | NA | 1210 | 0.160 | 0.4331 | Yes |
| 69 | FXN | NA | 1224 | 0.160 | 0.4376 | Yes |
| 70 | NDUFB6 | NA | 1229 | 0.159 | 0.4422 | Yes |
| 71 | NDUFA1 | NA | 1232 | 0.159 | 0.4468 | Yes |
| 72 | UQCRQ | NA | 1262 | 0.158 | 0.4509 | Yes |
| 73 | NDUFA4 | NA | 1317 | 0.156 | 0.4545 | Yes |
| 74 | SUCLG1 | NA | 1337 | 0.155 | 0.4586 | Yes |
| 75 | GPD1 | NA | 1419 | 0.151 | 0.4616 | Yes |
| 76 | ACADVL | NA | 1448 | 0.150 | 0.4655 | Yes |
| 77 | PARK7 | NA | 1571 | 0.146 | 0.4675 | Yes |
| 78 | NDUFA7 | NA | 1580 | 0.145 | 0.4716 | Yes |
| 79 | UQCR10 | NA | 1669 | 0.142 | 0.4741 | Yes |
| 80 | ETFB | NA | 1778 | 0.138 | 0.4762 | Yes |
| 81 | COQ9 | NA | 1858 | 0.135 | 0.4787 | Yes |
| 82 | CHCHD2 | NA | 1884 | 0.134 | 0.4822 | Yes |
| 83 | GYG2 | NA | 1994 | 0.131 | 0.4840 | Yes |
| 84 | GFPT2 | NA | 2022 | 0.130 | 0.4874 | Yes |
| 85 | NDUFB2 | NA | 2084 | 0.128 | 0.4900 | Yes |
| 86 | MT-CO1 | NA | 2118 | 0.127 | 0.4931 | Yes |
| 87 | NDUFV3 | NA | 2156 | 0.126 | 0.4962 | Yes |
| 88 | AKT2 | NA | 2199 | 0.125 | 0.4991 | Yes |
| 89 | PPP1R1A | NA | 2210 | 0.125 | 0.5026 | Yes |
| 90 | SDHA | NA | 2223 | 0.125 | 0.5060 | Yes |
| 91 | HIF1A | NA | 2230 | 0.125 | 0.5096 | Yes |
| 92 | TIGAR | NA | 2280 | 0.124 | 0.5123 | Yes |
| 93 | NDUFB3 | NA | 2295 | 0.124 | 0.5157 | Yes |
| 94 | PYGL | NA | 2341 | 0.123 | 0.5184 | Yes |
| 95 | GHITM | NA | 2408 | 0.121 | 0.5208 | Yes |
| 96 | PPP1R3C | NA | 2527 | 0.119 | 0.5221 | Yes |
| 97 | MT-ND6 | NA | 2559 | 0.118 | 0.5250 | Yes |
| 98 | NDUFS3 | NA | 2781 | 0.114 | 0.5242 | Yes |
| 99 | NDUFB8 | NA | 2814 | 0.113 | 0.5270 | Yes |
| 100 | PPP1CA | NA | 2906 | 0.111 | 0.5286 | Yes |
| 101 | COX6C | NA | 2950 | 0.111 | 0.5310 | Yes |
| 102 | FAHD1 | NA | 2966 | 0.110 | 0.5340 | Yes |
| 103 | MT-ND5 | NA | 2990 | 0.110 | 0.5368 | Yes |
| 104 | NDUFB11 | NA | 3141 | 0.107 | 0.5372 | Yes |
| 105 | NDUFA13 | NA | 3239 | 0.106 | 0.5385 | Yes |
| 106 | NDUFA12 | NA | 3245 | 0.106 | 0.5415 | Yes |
| 107 | SUCLA2 | NA | 3254 | 0.106 | 0.5444 | Yes |
| 108 | UGP2 | NA | 3327 | 0.104 | 0.5462 | Yes |
| 109 | POMC | NA | 3352 | 0.104 | 0.5488 | Yes |
| 110 | MT-ND4 | NA | 3412 | 0.103 | 0.5507 | Yes |
| 111 | COX4I1 | NA | 3492 | 0.102 | 0.5522 | Yes |
| 112 | MT-ND4L | NA | 3628 | 0.099 | 0.5527 | Yes |
| 113 | NDUFS8 | NA | 3741 | 0.098 | 0.5535 | Yes |
| 114 | UQCC3 | NA | 4344 | 0.089 | 0.5450 | Yes |
| 115 | IDH3B | NA | 4378 | 0.089 | 0.5470 | Yes |
| 116 | MT3 | NA | 4380 | 0.089 | 0.5496 | Yes |
| 117 | CAT | NA | 4420 | 0.088 | 0.5515 | Yes |
| 118 | CISD1 | NA | 4487 | 0.087 | 0.5528 | Yes |
| 119 | NDUFS7 | NA | 4492 | 0.087 | 0.5553 | Yes |
| 120 | UQCR11 | NA | 4792 | 0.084 | 0.5522 | No |
| 121 | NDUFA10 | NA | 4915 | 0.082 | 0.5524 | No |
| 122 | TP53 | NA | 5041 | 0.081 | 0.5525 | No |
| 123 | GSK3A | NA | 5504 | 0.075 | 0.5462 | No |
| 124 | BID | NA | 5583 | 0.074 | 0.5469 | No |
| 125 | SDHAF2 | NA | 5610 | 0.074 | 0.5486 | No |
| 126 | MDH1 | NA | 5615 | 0.074 | 0.5507 | No |
| 127 | MTFR1 | NA | 5966 | 0.070 | 0.5463 | No |
| 128 | SORBS1 | NA | 6060 | 0.069 | 0.5466 | No |
| 129 | SDHC | NA | 6151 | 0.068 | 0.5469 | No |
| 130 | NDUFA3 | NA | 6211 | 0.068 | 0.5478 | No |
| 131 | NDUFB10 | NA | 6353 | 0.066 | 0.5472 | No |
| 132 | MC4R | NA | 6389 | 0.066 | 0.5485 | No |
| 133 | NNT | NA | 6486 | 0.065 | 0.5486 | No |
| 134 | NDUFB7 | NA | 6634 | 0.064 | 0.5478 | No |
| 135 | GCK | NA | 6665 | 0.063 | 0.5491 | No |
| 136 | IRS2 | NA | 6918 | 0.061 | 0.5462 | No |
| 137 | OGDHL | NA | 7326 | 0.058 | 0.5404 | No |
| 138 | PYGB | NA | 7737 | 0.054 | 0.5345 | No |
| 139 | IDH3G | NA | 7850 | 0.053 | 0.5340 | No |
| 140 | PDHA2 | NA | 7891 | 0.053 | 0.5348 | No |
| 141 | NDUFB1 | NA | 8300 | 0.049 | 0.5287 | No |
| 142 | MYC | NA | 8467 | 0.048 | 0.5271 | No |
| 143 | SDHD | NA | 8747 | 0.046 | 0.5233 | No |
| 144 | OPN3 | NA | 8879 | 0.045 | 0.5222 | No |
| 145 | NDUFA2 | NA | 9287 | 0.042 | 0.5159 | No |
| 146 | IDH1 | NA | 9327 | 0.042 | 0.5164 | No |
| 147 | CCNB1 | NA | 9393 | 0.041 | 0.5164 | No |
| 148 | COQ10B | NA | 10565 | 0.033 | 0.4958 | No |
| 149 | NDUFS4 | NA | 10698 | 0.032 | 0.4944 | No |
| 150 | MT-CO2 | NA | 11073 | 0.030 | 0.4884 | No |
| 151 | BNIP3 | NA | 11495 | 0.027 | 0.4814 | No |
| 152 | SLC25A25 | NA | 11707 | 0.026 | 0.4783 | No |
| 153 | NR4A3 | NA | 12046 | 0.024 | 0.4728 | No |
| 154 | DHTKD1 | NA | 12265 | 0.023 | 0.4694 | No |
| 155 | DNAJC15 | NA | 12342 | 0.022 | 0.4687 | No |
| 156 | MTCO2P12 | NA | 12469 | 0.022 | 0.4670 | No |
| 157 | DGUOK | NA | 12697 | 0.020 | 0.4634 | No |
| 158 | ME2 | NA | 12983 | 0.019 | 0.4587 | No |
| 159 | PHKA2 | NA | 12991 | 0.019 | 0.4591 | No |
| 160 | PRLH | NA | 13574 | 0.016 | 0.4489 | No |
| 161 | ACSM1 | NA | 13801 | 0.015 | 0.4451 | No |
| 162 | PGM2 | NA | 13917 | 0.014 | 0.4435 | No |
| 163 | PER2 | NA | 14933 | 0.010 | 0.4250 | No |
| 164 | NDUFB4 | NA | 15916 | 0.006 | 0.4071 | No |
| 165 | GYG1 | NA | 16076 | 0.005 | 0.4044 | No |
| 166 | NDUFB5 | NA | 16097 | 0.005 | 0.4041 | No |
| 167 | SUCLG2 | NA | 17313 | 0.000 | 0.3818 | No |
| 168 | GSK3B | NA | 17338 | 0.000 | 0.3813 | No |
| 169 | UQCRB | NA | 21357 | -0.000 | 0.3074 | No |
| 170 | GPD2 | NA | 21399 | -0.000 | 0.3066 | No |
| 171 | MT-CO3 | NA | 21694 | -0.002 | 0.3013 | No |
| 172 | MTOR | NA | 23776 | -0.008 | 0.2632 | No |
| 173 | INPP5K | NA | 24263 | -0.010 | 0.2545 | No |
| 174 | GNAS | NA | 24861 | -0.012 | 0.2439 | No |
| 175 | NDUFV1 | NA | 25399 | -0.014 | 0.2344 | No |
| 176 | PPP1R2P1 | NA | 25585 | -0.015 | 0.2315 | No |
| 177 | NHLRC1 | NA | 26660 | -0.020 | 0.2123 | No |
| 178 | NDUFC1 | NA | 26958 | -0.021 | 0.2074 | No |
| 179 | BLOC1S1 | NA | 27753 | -0.024 | 0.1935 | No |
| 180 | COX15 | NA | 28194 | -0.027 | 0.1862 | No |
| 181 | SLC25A23 | NA | 28199 | -0.027 | 0.1869 | No |
| 182 | NOA1 | NA | 28680 | -0.029 | 0.1789 | No |
| 183 | NDUFA11 | NA | 28809 | -0.029 | 0.1774 | No |
| 184 | RUBCNL | NA | 29736 | -0.034 | 0.1614 | No |
| 185 | PHKG2 | NA | 30163 | -0.036 | 0.1546 | No |
| 186 | CALM1 | NA | 30883 | -0.039 | 0.1425 | No |
| 187 | PINK1 | NA | 30921 | -0.039 | 0.1429 | No |
| 188 | CDK1 | NA | 31040 | -0.040 | 0.1419 | No |
| 189 | NDUFA5 | NA | 31427 | -0.042 | 0.1361 | No |
| 190 | MDH1B | NA | 31580 | -0.042 | 0.1345 | No |
| 191 | OXA1L | NA | 31784 | -0.043 | 0.1320 | No |
| 192 | PIK3CA | NA | 31899 | -0.044 | 0.1312 | No |
| 193 | IL4 | NA | 32256 | -0.046 | 0.1260 | No |
| 194 | GCGR | NA | 32496 | -0.047 | 0.1230 | No |
| 195 | MTFR2 | NA | 33001 | -0.049 | 0.1152 | No |
| 196 | ME3 | NA | 33187 | -0.050 | 0.1132 | No |
| 197 | MT-CYB | NA | 33330 | -0.051 | 0.1121 | No |
| 198 | MIR210 | NA | 33341 | -0.051 | 0.1135 | No |
| 199 | PGM2L1 | NA | 33415 | -0.052 | 0.1136 | No |
| 200 | PRKAG2 | NA | 33741 | -0.053 | 0.1092 | No |
| 201 | MT-ND1 | NA | 34219 | -0.056 | 0.1021 | No |
| 202 | SLC25A18 | NA | 34848 | -0.059 | 0.0922 | No |
| 203 | PHKG1 | NA | 34886 | -0.060 | 0.0933 | No |
| 204 | PANK2 | NA | 34973 | -0.060 | 0.0935 | No |
| 205 | PPP1CC | NA | 35641 | -0.064 | 0.0831 | No |
| 206 | NR1D1 | NA | 36186 | -0.067 | 0.0750 | No |
| 207 | COX6A2 | NA | 36212 | -0.067 | 0.0765 | No |
| 208 | ETFRF1 | NA | 36529 | -0.068 | 0.0727 | No |
| 209 | PRDM16 | NA | 36622 | -0.069 | 0.0730 | No |
| 210 | SLC25A13 | NA | 37233 | -0.072 | 0.0639 | No |
| 211 | INS | NA | 37337 | -0.073 | 0.0641 | No |
| 212 | PCDH12 | NA | 37416 | -0.073 | 0.0649 | No |
| 213 | INSR | NA | 37534 | -0.074 | 0.0649 | No |
| 214 | MTFR1L | NA | 37742 | -0.075 | 0.0632 | No |
| 215 | G6PC | NA | 37746 | -0.075 | 0.0654 | No |
| 216 | COX7A2L | NA | 37835 | -0.075 | 0.0660 | No |
| 217 | PHKA1 | NA | 37931 | -0.076 | 0.0664 | No |
| 218 | PPP1R3A | NA | 37982 | -0.076 | 0.0678 | No |
| 219 | IDE | NA | 38060 | -0.077 | 0.0686 | No |
| 220 | UCN | NA | 38063 | -0.077 | 0.0708 | No |
| 221 | LEP | NA | 38312 | -0.078 | 0.0685 | No |
| 222 | PPP1R3F | NA | 38479 | -0.079 | 0.0678 | No |
| 223 | SURF1 | NA | 38823 | -0.081 | 0.0638 | No |
| 224 | SNCA | NA | 39076 | -0.083 | 0.0616 | No |
| 225 | SDHAF4 | NA | 40046 | -0.088 | 0.0464 | No |
| 226 | COX4I2 | NA | 40080 | -0.089 | 0.0484 | No |
| 227 | IMMP2L | NA | 40105 | -0.089 | 0.0505 | No |
| 228 | IL6ST | NA | 40622 | -0.092 | 0.0437 | No |
| 229 | PTH | NA | 40626 | -0.092 | 0.0464 | No |
| 230 | IREB2 | NA | 40994 | -0.094 | 0.0424 | No |
| 231 | NFATC4 | NA | 41191 | -0.095 | 0.0415 | No |
| 232 | NOS2 | NA | 42122 | -0.101 | 0.0274 | No |
| 233 | ACTN3 | NA | 42391 | -0.103 | 0.0255 | No |
| 234 | GNMT | NA | 42406 | -0.103 | 0.0282 | No |
| 235 | LYRM7 | NA | 42432 | -0.103 | 0.0308 | No |
| 236 | PRKAG3 | NA | 42933 | -0.106 | 0.0247 | No |
| 237 | MT-ND3 | NA | 43070 | -0.107 | 0.0254 | No |
| 238 | KHK | NA | 43862 | -0.113 | 0.0141 | No |
| 239 | GYS2 | NA | 44049 | -0.114 | 0.0140 | No |
| 240 | AGL | NA | 44690 | -0.119 | 0.0057 | No |
| 241 | PPARGC1A | NA | 45150 | -0.122 | 0.0009 | No |
| 242 | PID1 | NA | 45478 | -0.125 | -0.0015 | No |
| 243 | IGF2 | NA | 46221 | -0.130 | -0.0113 | No |
| 244 | ENPP1 | NA | 46658 | -0.134 | -0.0154 | No |
| 245 | PPP1R3B | NA | 46914 | -0.136 | -0.0161 | No |
| 246 | COQ10A | NA | 47189 | -0.139 | -0.0171 | No |
| 247 | KL | NA | 47199 | -0.139 | -0.0132 | No |
| 248 | PASK | NA | 47369 | -0.140 | -0.0122 | No |
| 249 | PPP1R2 | NA | 47392 | -0.140 | -0.0085 | No |
| 250 | EPM2AIP1 | NA | 47492 | -0.141 | -0.0061 | No |
| 251 | ESRRB | NA | 47807 | -0.144 | -0.0077 | No |
| 252 | PPP1R3G | NA | 49009 | -0.155 | -0.0253 | No |
| 253 | PPP1R3D | NA | 49262 | -0.157 | -0.0253 | No |
| 254 | HMGB1 | NA | 49773 | -0.163 | -0.0299 | No |
| 255 | DYRK2 | NA | 50199 | -0.167 | -0.0328 | No |
| 256 | SIRT3 | NA | 50705 | -0.173 | -0.0370 | No |
| 257 | RB1CC1 | NA | 50745 | -0.174 | -0.0327 | No |
| 258 | SLC25A14 | NA | 51238 | -0.180 | -0.0364 | No |
| 259 | MRAP2 | NA | 51388 | -0.182 | -0.0338 | No |
| 260 | ISCU | NA | 51531 | -0.184 | -0.0310 | No |
| 261 | POLG2 | NA | 52404 | -0.199 | -0.0413 | No |
| 262 | IFNG | NA | 52552 | -0.201 | -0.0381 | No |
| 263 | PHKB | NA | 52653 | -0.203 | -0.0339 | No |
| 264 | TREX1 | NA | 52834 | -0.207 | -0.0312 | No |
| 265 | SLC25A12 | NA | 52897 | -0.208 | -0.0262 | No |
| 266 | PPP1CB | NA | 52905 | -0.209 | -0.0202 | No |
| 267 | ADGRF5 | NA | 53295 | -0.218 | -0.0210 | No |
| 268 | LEPR | NA | 53544 | -0.226 | -0.0189 | No |
| 269 | GAA | NA | 53612 | -0.228 | -0.0135 | No |
| 270 | MT-ND2 | NA | 53712 | -0.232 | -0.0085 | No |
| 271 | PPP1R3E | NA | 54296 | -0.262 | -0.0115 | No |
| 272 | TAZ | NA | 54454 | -0.280 | -0.0062 | No |
| 273 | CBFA2T3 | NA | 54530 | -0.297 | 0.0011 | No |