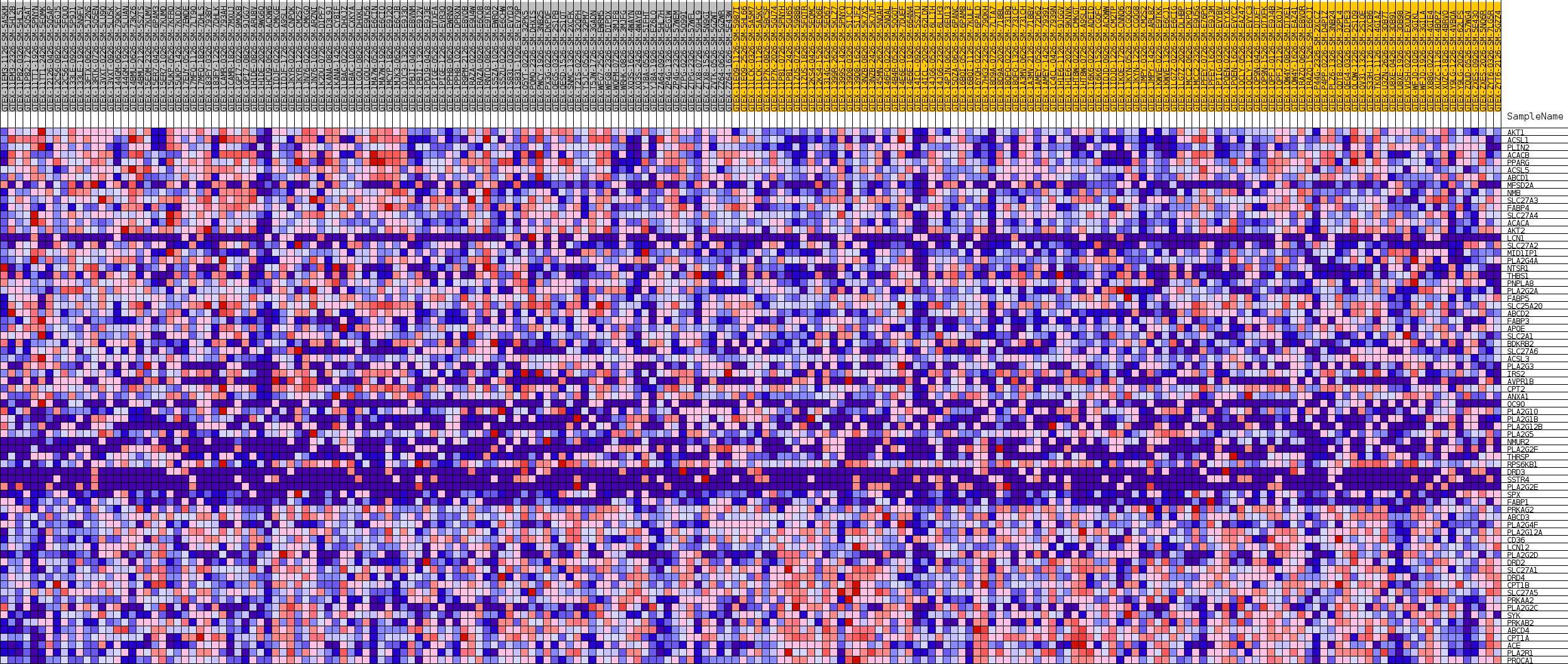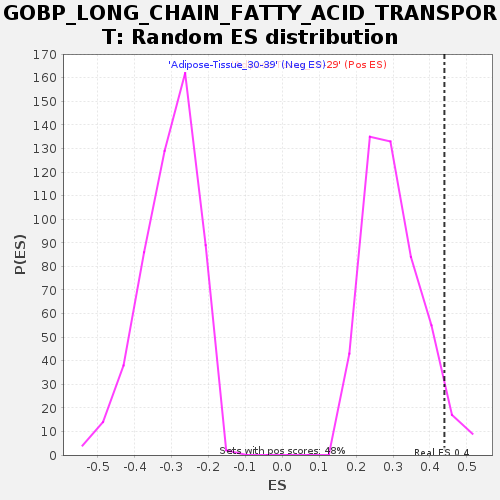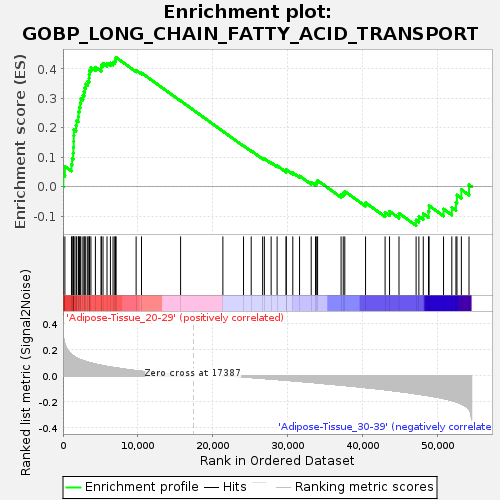
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_20-29 |
| GeneSet | GOBP_LONG_CHAIN_FATTY_ACID_TRANSPORT |
| Enrichment Score (ES) | 0.4391041 |
| Normalized Enrichment Score (NES) | 1.463637 |
| Nominal p-value | 0.04621849 |
| FDR q-value | 0.5463713 |
| FWER p-Value | 0.97 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | AKT1 | NA | 49 | 0.299 | 0.0397 | Yes |
| 2 | ACSL1 | NA | 252 | 0.244 | 0.0691 | Yes |
| 3 | PLIN2 | NA | 1130 | 0.164 | 0.0752 | Yes |
| 4 | ACACB | NA | 1216 | 0.160 | 0.0954 | Yes |
| 5 | PPARG | NA | 1353 | 0.154 | 0.1137 | Yes |
| 6 | ACSL5 | NA | 1384 | 0.153 | 0.1339 | Yes |
| 7 | ABCD1 | NA | 1427 | 0.151 | 0.1536 | Yes |
| 8 | MFSD2A | NA | 1435 | 0.151 | 0.1739 | Yes |
| 9 | NMB | NA | 1460 | 0.150 | 0.1938 | Yes |
| 10 | SLC27A3 | NA | 1738 | 0.139 | 0.2075 | Yes |
| 11 | FABP4 | NA | 1824 | 0.136 | 0.2245 | Yes |
| 12 | SLC27A4 | NA | 2050 | 0.129 | 0.2379 | Yes |
| 13 | ACACA | NA | 2079 | 0.129 | 0.2548 | Yes |
| 14 | AKT2 | NA | 2199 | 0.125 | 0.2696 | Yes |
| 15 | LCN1 | NA | 2302 | 0.123 | 0.2845 | Yes |
| 16 | SLC27A2 | NA | 2381 | 0.122 | 0.2995 | Yes |
| 17 | MID1IP1 | NA | 2660 | 0.116 | 0.3102 | Yes |
| 18 | PLA2G4A | NA | 2812 | 0.113 | 0.3228 | Yes |
| 19 | NTSR1 | NA | 2913 | 0.111 | 0.3360 | Yes |
| 20 | THBS1 | NA | 3043 | 0.109 | 0.3485 | Yes |
| 21 | PNPLA8 | NA | 3300 | 0.105 | 0.3580 | Yes |
| 22 | PLA2G2A | NA | 3479 | 0.102 | 0.3685 | Yes |
| 23 | FABP5 | NA | 3494 | 0.102 | 0.3820 | Yes |
| 24 | SLC25A20 | NA | 3546 | 0.101 | 0.3948 | Yes |
| 25 | ABCD2 | NA | 3729 | 0.098 | 0.4047 | Yes |
| 26 | FABP3 | NA | 4330 | 0.089 | 0.4058 | Yes |
| 27 | APOE | NA | 5094 | 0.080 | 0.4026 | Yes |
| 28 | SLC2A1 | NA | 5118 | 0.080 | 0.4130 | Yes |
| 29 | BDKRB2 | NA | 5348 | 0.077 | 0.4193 | Yes |
| 30 | SLC27A6 | NA | 5881 | 0.071 | 0.4191 | Yes |
| 31 | ACSL3 | NA | 6336 | 0.066 | 0.4198 | Yes |
| 32 | PLA2G3 | NA | 6686 | 0.063 | 0.4220 | Yes |
| 33 | IRS2 | NA | 6918 | 0.061 | 0.4260 | Yes |
| 34 | AVPR1B | NA | 7013 | 0.060 | 0.4325 | Yes |
| 35 | CPT2 | NA | 7094 | 0.060 | 0.4391 | Yes |
| 36 | ANXA1 | NA | 9769 | 0.039 | 0.3953 | No |
| 37 | OC90 | NA | 10489 | 0.034 | 0.3867 | No |
| 38 | PLA2G10 | NA | 15712 | 0.006 | 0.2918 | No |
| 39 | PLA2G1B | NA | 21362 | -0.000 | 0.1882 | No |
| 40 | PLA2G12B | NA | 24121 | -0.009 | 0.1388 | No |
| 41 | PLA2G5 | NA | 25146 | -0.013 | 0.1219 | No |
| 42 | NMUR2 | NA | 26676 | -0.020 | 0.0965 | No |
| 43 | PLA2G2F | NA | 26905 | -0.021 | 0.0951 | No |
| 44 | THRSP | NA | 27811 | -0.025 | 0.0819 | No |
| 45 | RPS6KB1 | NA | 28606 | -0.029 | 0.0712 | No |
| 46 | DRD3 | NA | 29808 | -0.034 | 0.0538 | No |
| 47 | SSTR4 | NA | 29829 | -0.034 | 0.0580 | No |
| 48 | PLA2G2E | NA | 30705 | -0.038 | 0.0472 | No |
| 49 | SPX | NA | 31606 | -0.043 | 0.0364 | No |
| 50 | FABP1 | NA | 33163 | -0.050 | 0.0147 | No |
| 51 | PRKAG2 | NA | 33741 | -0.053 | 0.0113 | No |
| 52 | ABCD3 | NA | 33906 | -0.054 | 0.0157 | No |
| 53 | PLA2G4F | NA | 34015 | -0.055 | 0.0211 | No |
| 54 | PLA2G12A | NA | 37154 | -0.072 | -0.0267 | No |
| 55 | CD36 | NA | 37443 | -0.073 | -0.0220 | No |
| 56 | LCN12 | NA | 37651 | -0.074 | -0.0158 | No |
| 57 | PLA2G2D | NA | 40425 | -0.091 | -0.0544 | No |
| 58 | DRD2 | NA | 43036 | -0.107 | -0.0877 | No |
| 59 | SLC27A1 | NA | 43630 | -0.111 | -0.0835 | No |
| 60 | DRD4 | NA | 44894 | -0.120 | -0.0904 | No |
| 61 | CPT1B | NA | 47179 | -0.138 | -0.1135 | No |
| 62 | SLC27A5 | NA | 47551 | -0.142 | -0.1011 | No |
| 63 | PRKAA2 | NA | 48137 | -0.147 | -0.0919 | No |
| 64 | PLA2G2C | NA | 48838 | -0.153 | -0.0839 | No |
| 65 | SYK | NA | 48916 | -0.154 | -0.0645 | No |
| 66 | PRKAB2 | NA | 50845 | -0.175 | -0.0761 | No |
| 67 | ABCD4 | NA | 51951 | -0.191 | -0.0705 | No |
| 68 | CPT1A | NA | 52503 | -0.200 | -0.0535 | No |
| 69 | ACE | NA | 52637 | -0.203 | -0.0284 | No |
| 70 | PLA2R1 | NA | 53228 | -0.216 | -0.0099 | No |
| 71 | PROCA1 | NA | 54248 | -0.257 | 0.0063 | No |