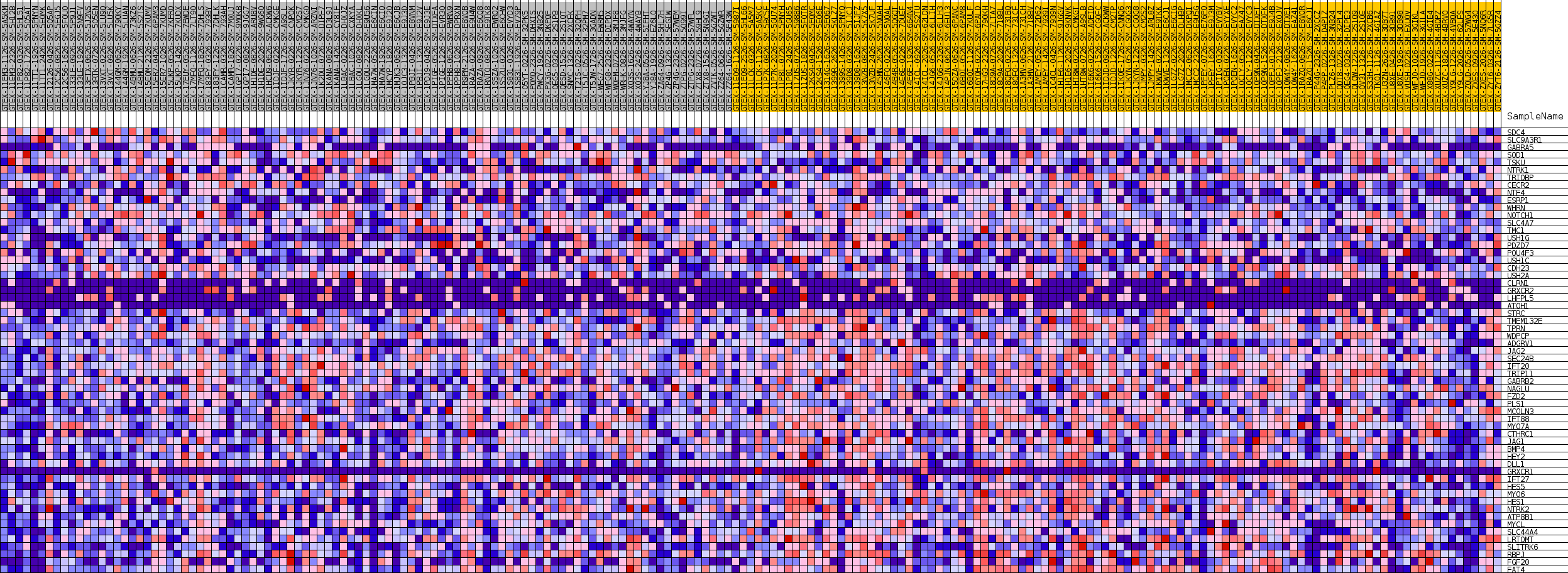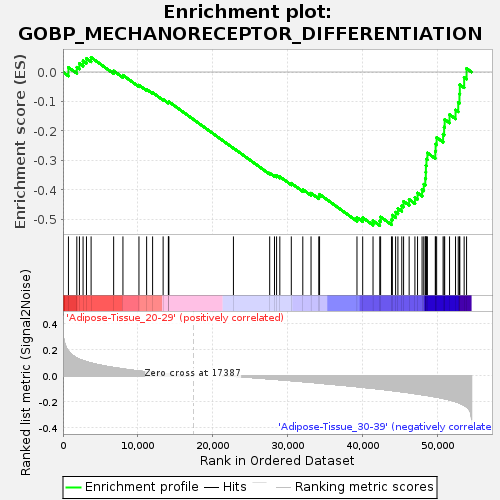
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_30-39 |
| GeneSet | GOBP_MECHANORECEPTOR_DIFFERENTIATION |
| Enrichment Score (ES) | -0.5213389 |
| Normalized Enrichment Score (NES) | -1.6421952 |
| Nominal p-value | 0.008080808 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.833 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SDC4 | NA | 727 | 0.190 | 0.0158 | No |
| 2 | SLC9A3R1 | NA | 1846 | 0.136 | 0.0161 | No |
| 3 | GABRA5 | NA | 2196 | 0.126 | 0.0290 | No |
| 4 | SOD1 | NA | 2681 | 0.116 | 0.0378 | No |
| 5 | TSKU | NA | 3136 | 0.107 | 0.0459 | No |
| 6 | NTRK1 | NA | 3755 | 0.098 | 0.0496 | No |
| 7 | TRIOBP | NA | 6767 | 0.062 | 0.0039 | No |
| 8 | CECR2 | NA | 8008 | 0.052 | -0.0109 | No |
| 9 | NTF4 | NA | 10148 | 0.036 | -0.0446 | No |
| 10 | ESRP1 | NA | 11174 | 0.029 | -0.0590 | No |
| 11 | WHRN | NA | 11972 | 0.025 | -0.0698 | No |
| 12 | NOTCH1 | NA | 13378 | 0.017 | -0.0930 | No |
| 13 | SLC4A7 | NA | 14085 | 0.014 | -0.1038 | No |
| 14 | TMC1 | NA | 14087 | 0.014 | -0.1018 | No |
| 15 | USH1G | NA | 14146 | 0.013 | -0.1008 | No |
| 16 | PDZD7 | NA | 22772 | -0.005 | -0.2583 | No |
| 17 | POU4F3 | NA | 27616 | -0.024 | -0.3434 | No |
| 18 | USH1C | NA | 28257 | -0.027 | -0.3510 | No |
| 19 | CDH23 | NA | 28538 | -0.028 | -0.3518 | No |
| 20 | USH2A | NA | 28970 | -0.030 | -0.3551 | No |
| 21 | CLRN1 | NA | 30504 | -0.037 | -0.3775 | No |
| 22 | GRXCR2 | NA | 32040 | -0.045 | -0.3988 | No |
| 23 | LHFPL5 | NA | 33142 | -0.050 | -0.4113 | No |
| 24 | ATOH1 | NA | 34176 | -0.056 | -0.4217 | No |
| 25 | STRC | NA | 34281 | -0.056 | -0.4150 | No |
| 26 | TMEM132E | NA | 39279 | -0.084 | -0.4938 | No |
| 27 | TPRN | NA | 40043 | -0.088 | -0.4943 | No |
| 28 | WDPCP | NA | 41424 | -0.097 | -0.5048 | No |
| 29 | ADGRV1 | NA | 42326 | -0.103 | -0.5056 | Yes |
| 30 | JAG2 | NA | 42438 | -0.103 | -0.4919 | Yes |
| 31 | SEC24B | NA | 43889 | -0.113 | -0.5011 | Yes |
| 32 | IFT20 | NA | 44009 | -0.114 | -0.4859 | Yes |
| 33 | TRIP11 | NA | 44441 | -0.117 | -0.4759 | Yes |
| 34 | GABRB2 | NA | 44748 | -0.119 | -0.4632 | Yes |
| 35 | NAGLU | NA | 45266 | -0.123 | -0.4538 | Yes |
| 36 | FZD2 | NA | 45508 | -0.125 | -0.4391 | Yes |
| 37 | PLS1 | NA | 46250 | -0.131 | -0.4326 | Yes |
| 38 | MCOLN3 | NA | 47020 | -0.137 | -0.4257 | Yes |
| 39 | IFT88 | NA | 47372 | -0.140 | -0.4107 | Yes |
| 40 | MYO7A | NA | 47977 | -0.146 | -0.3994 | Yes |
| 41 | CTHRC1 | NA | 48204 | -0.148 | -0.3809 | Yes |
| 42 | JAG1 | NA | 48404 | -0.149 | -0.3617 | Yes |
| 43 | BMP4 | NA | 48475 | -0.150 | -0.3400 | Yes |
| 44 | HEY2 | NA | 48488 | -0.150 | -0.3172 | Yes |
| 45 | DLL1 | NA | 48561 | -0.151 | -0.2955 | Yes |
| 46 | GRXCR1 | NA | 48689 | -0.152 | -0.2745 | Yes |
| 47 | IFT27 | NA | 49744 | -0.162 | -0.2690 | Yes |
| 48 | HES5 | NA | 49786 | -0.163 | -0.2448 | Yes |
| 49 | MYO6 | NA | 49924 | -0.164 | -0.2222 | Yes |
| 50 | HES1 | NA | 50785 | -0.174 | -0.2112 | Yes |
| 51 | NTRK2 | NA | 50923 | -0.176 | -0.1868 | Yes |
| 52 | ATP8B1 | NA | 50998 | -0.177 | -0.1610 | Yes |
| 53 | MYCL | NA | 51647 | -0.186 | -0.1444 | Yes |
| 54 | SLC44A4 | NA | 52448 | -0.199 | -0.1286 | Yes |
| 55 | LRTOMT | NA | 52810 | -0.206 | -0.1036 | Yes |
| 56 | SLITRK6 | NA | 52972 | -0.210 | -0.0743 | Yes |
| 57 | RBPJ | NA | 53022 | -0.211 | -0.0428 | Yes |
| 58 | FGF20 | NA | 53600 | -0.228 | -0.0185 | Yes |
| 59 | FAT4 | NA | 53923 | -0.239 | 0.0122 | Yes |