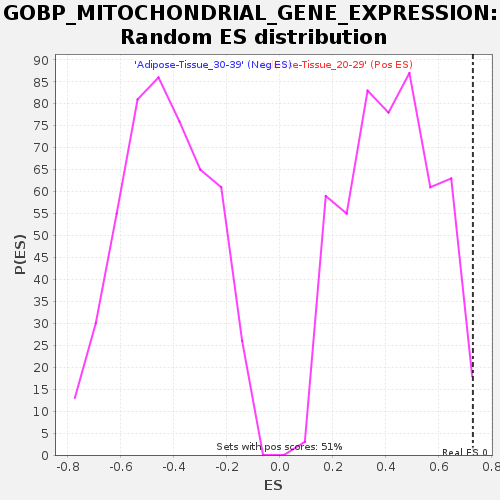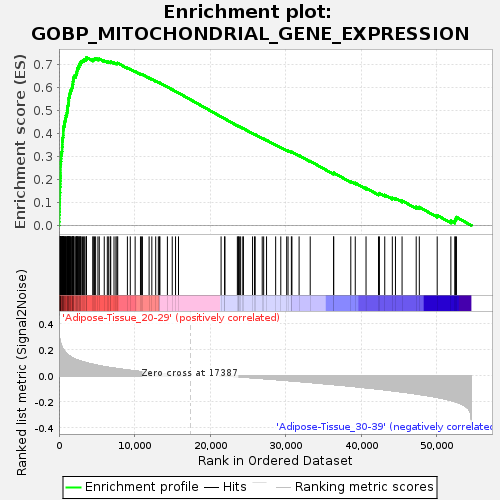
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_20-29 |
| GeneSet | GOBP_MITOCHONDRIAL_GENE_EXPRESSION |
| Enrichment Score (ES) | 0.7296393 |
| Normalized Enrichment Score (NES) | 1.7124463 |
| Nominal p-value | 0.007889546 |
| FDR q-value | 0.21638912 |
| FWER p-Value | 0.692 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | C1QBP | NA | 19 | 0.332 | 0.0159 | Yes |
| 2 | MRPS18A | NA | 31 | 0.314 | 0.0310 | Yes |
| 3 | MRPL14 | NA | 34 | 0.311 | 0.0462 | Yes |
| 4 | TSFM | NA | 63 | 0.294 | 0.0601 | Yes |
| 5 | MRPL36 | NA | 79 | 0.287 | 0.0738 | Yes |
| 6 | MRPS11 | NA | 82 | 0.285 | 0.0877 | Yes |
| 7 | MALSU1 | NA | 85 | 0.284 | 0.1015 | Yes |
| 8 | HSD17B10 | NA | 97 | 0.280 | 0.1150 | Yes |
| 9 | MRPL46 | NA | 108 | 0.277 | 0.1284 | Yes |
| 10 | MRPS2 | NA | 117 | 0.274 | 0.1416 | Yes |
| 11 | MRPS7 | NA | 142 | 0.266 | 0.1542 | Yes |
| 12 | MRPL4 | NA | 143 | 0.266 | 0.1672 | Yes |
| 13 | MRPS18C | NA | 164 | 0.259 | 0.1795 | Yes |
| 14 | TUFM | NA | 169 | 0.259 | 0.1921 | Yes |
| 15 | MRPL15 | NA | 178 | 0.257 | 0.2045 | Yes |
| 16 | MRPL12 | NA | 189 | 0.255 | 0.2168 | Yes |
| 17 | MRPL17 | NA | 202 | 0.253 | 0.2289 | Yes |
| 18 | TRUB2 | NA | 204 | 0.253 | 0.2413 | Yes |
| 19 | CHCHD10 | NA | 205 | 0.253 | 0.2536 | Yes |
| 20 | MRPS16 | NA | 208 | 0.252 | 0.2659 | Yes |
| 21 | MRPS17 | NA | 242 | 0.246 | 0.2773 | Yes |
| 22 | MRPS35 | NA | 269 | 0.242 | 0.2887 | Yes |
| 23 | MRPL3 | NA | 280 | 0.240 | 0.3002 | Yes |
| 24 | MRPL58 | NA | 299 | 0.237 | 0.3114 | Yes |
| 25 | MRPS22 | NA | 373 | 0.224 | 0.3211 | Yes |
| 26 | YARS2 | NA | 419 | 0.218 | 0.3309 | Yes |
| 27 | MRPL24 | NA | 421 | 0.218 | 0.3415 | Yes |
| 28 | MRPL37 | NA | 445 | 0.216 | 0.3517 | Yes |
| 29 | PUS1 | NA | 448 | 0.215 | 0.3621 | Yes |
| 30 | MRPS12 | NA | 454 | 0.214 | 0.3725 | Yes |
| 31 | TBRG4 | NA | 492 | 0.210 | 0.3821 | Yes |
| 32 | MRPL20 | NA | 556 | 0.204 | 0.3909 | Yes |
| 33 | MRPS23 | NA | 565 | 0.203 | 0.4007 | Yes |
| 34 | MRPS28 | NA | 567 | 0.203 | 0.4106 | Yes |
| 35 | MRPL47 | NA | 573 | 0.202 | 0.4203 | Yes |
| 36 | MPV17L2 | NA | 599 | 0.200 | 0.4297 | Yes |
| 37 | TWNK | NA | 704 | 0.192 | 0.4371 | Yes |
| 38 | MRPL2 | NA | 731 | 0.190 | 0.4459 | Yes |
| 39 | PPARGC1B | NA | 781 | 0.186 | 0.4542 | Yes |
| 40 | GFM1 | NA | 861 | 0.181 | 0.4615 | Yes |
| 41 | SHMT2 | NA | 897 | 0.179 | 0.4696 | Yes |
| 42 | PNPT1 | NA | 923 | 0.177 | 0.4778 | Yes |
| 43 | MRPL27 | NA | 1036 | 0.170 | 0.4840 | Yes |
| 44 | MRPL21 | NA | 1061 | 0.168 | 0.4918 | Yes |
| 45 | MRPS5 | NA | 1080 | 0.167 | 0.4996 | Yes |
| 46 | TFB2M | NA | 1128 | 0.164 | 0.5068 | Yes |
| 47 | MRPL32 | NA | 1151 | 0.163 | 0.5143 | Yes |
| 48 | FASTKD2 | NA | 1175 | 0.162 | 0.5219 | Yes |
| 49 | MRPL19 | NA | 1244 | 0.158 | 0.5283 | Yes |
| 50 | TRMT10C | NA | 1286 | 0.157 | 0.5352 | Yes |
| 51 | MRPL35 | NA | 1298 | 0.156 | 0.5427 | Yes |
| 52 | AURKAIP1 | NA | 1304 | 0.156 | 0.5502 | Yes |
| 53 | CHCHD1 | NA | 1326 | 0.155 | 0.5574 | Yes |
| 54 | DARS2 | NA | 1403 | 0.152 | 0.5634 | Yes |
| 55 | SLC25A33 | NA | 1416 | 0.152 | 0.5706 | Yes |
| 56 | GADD45GIP1 | NA | 1470 | 0.149 | 0.5770 | Yes |
| 57 | MRPL50 | NA | 1512 | 0.148 | 0.5834 | Yes |
| 58 | NDUFA7 | NA | 1580 | 0.145 | 0.5893 | Yes |
| 59 | LRPPRC | NA | 1684 | 0.141 | 0.5943 | Yes |
| 60 | ERAL1 | NA | 1736 | 0.139 | 0.6002 | Yes |
| 61 | TFAM | NA | 1772 | 0.138 | 0.6063 | Yes |
| 62 | MRPS15 | NA | 1816 | 0.137 | 0.6122 | Yes |
| 63 | ELAC2 | NA | 1847 | 0.136 | 0.6183 | Yes |
| 64 | MRPL38 | NA | 1855 | 0.135 | 0.6248 | Yes |
| 65 | MRPS33 | NA | 1883 | 0.134 | 0.6308 | Yes |
| 66 | MRPL39 | NA | 1885 | 0.134 | 0.6374 | Yes |
| 67 | NGRN | NA | 1971 | 0.132 | 0.6423 | Yes |
| 68 | SUPV3L1 | NA | 1993 | 0.131 | 0.6483 | Yes |
| 69 | MRPS36 | NA | 2226 | 0.125 | 0.6501 | Yes |
| 70 | GATB | NA | 2246 | 0.125 | 0.6559 | Yes |
| 71 | MRPL13 | NA | 2312 | 0.123 | 0.6607 | Yes |
| 72 | MRPS18B | NA | 2366 | 0.122 | 0.6657 | Yes |
| 73 | EARS2 | NA | 2382 | 0.122 | 0.6714 | Yes |
| 74 | TACO1 | NA | 2417 | 0.121 | 0.6766 | Yes |
| 75 | DAP3 | NA | 2421 | 0.121 | 0.6825 | Yes |
| 76 | MRPS26 | NA | 2553 | 0.118 | 0.6859 | Yes |
| 77 | MRPL57 | NA | 2651 | 0.116 | 0.6898 | Yes |
| 78 | MTG1 | NA | 2652 | 0.116 | 0.6955 | Yes |
| 79 | FASTKD5 | NA | 2688 | 0.116 | 0.7005 | Yes |
| 80 | MRPL44 | NA | 2866 | 0.112 | 0.7027 | Yes |
| 81 | MRPL22 | NA | 2884 | 0.112 | 0.7078 | Yes |
| 82 | MRPS34 | NA | 2912 | 0.111 | 0.7128 | Yes |
| 83 | MRPL52 | NA | 3105 | 0.108 | 0.7145 | Yes |
| 84 | IARS2 | NA | 3259 | 0.105 | 0.7169 | Yes |
| 85 | GFM2 | NA | 3355 | 0.104 | 0.7202 | Yes |
| 86 | MRPL16 | NA | 3577 | 0.100 | 0.7211 | Yes |
| 87 | MRPS24 | NA | 3620 | 0.099 | 0.7251 | Yes |
| 88 | TRNT1 | NA | 3640 | 0.099 | 0.7296 | Yes |
| 89 | MRPL9 | NA | 4471 | 0.088 | 0.7187 | No |
| 90 | COA3 | NA | 4540 | 0.087 | 0.7217 | No |
| 91 | MRPS9 | NA | 4693 | 0.085 | 0.7230 | No |
| 92 | RCC1L | NA | 4847 | 0.083 | 0.7242 | No |
| 93 | UQCC2 | NA | 5154 | 0.079 | 0.7225 | No |
| 94 | RPUSD3 | NA | 5331 | 0.077 | 0.7230 | No |
| 95 | UQCC1 | NA | 5990 | 0.070 | 0.7144 | No |
| 96 | LARS2 | NA | 6397 | 0.066 | 0.7101 | No |
| 97 | MRPL54 | NA | 6576 | 0.064 | 0.7100 | No |
| 98 | RPUSD4 | NA | 6792 | 0.062 | 0.7091 | No |
| 99 | MRPL51 | NA | 6849 | 0.062 | 0.7111 | No |
| 100 | MRPS10 | NA | 7281 | 0.058 | 0.7060 | No |
| 101 | MRPL40 | NA | 7535 | 0.056 | 0.7041 | No |
| 102 | MRPS30 | NA | 7675 | 0.055 | 0.7042 | No |
| 103 | TRMT5 | NA | 7773 | 0.054 | 0.7050 | No |
| 104 | TRMT61B | NA | 9054 | 0.044 | 0.6836 | No |
| 105 | MRPL30 | NA | 9438 | 0.041 | 0.6786 | No |
| 106 | MRPL11 | NA | 10080 | 0.037 | 0.6686 | No |
| 107 | MRPL41 | NA | 10784 | 0.032 | 0.6572 | No |
| 108 | MRPL28 | NA | 10908 | 0.031 | 0.6565 | No |
| 109 | MRPL18 | NA | 11044 | 0.030 | 0.6555 | No |
| 110 | GATC | NA | 11928 | 0.025 | 0.6405 | No |
| 111 | POLRMT | NA | 12289 | 0.023 | 0.6350 | No |
| 112 | MRPL23 | NA | 12802 | 0.020 | 0.6265 | No |
| 113 | MRPS31 | NA | 13151 | 0.018 | 0.6210 | No |
| 114 | MRPL1 | NA | 13319 | 0.017 | 0.6188 | No |
| 115 | FASTKD3 | NA | 13352 | 0.017 | 0.6190 | No |
| 116 | PTCD3 | NA | 14349 | 0.012 | 0.6013 | No |
| 117 | TRIT1 | NA | 14991 | 0.009 | 0.5900 | No |
| 118 | TARS2 | NA | 15427 | 0.007 | 0.5824 | No |
| 119 | MRPL10 | NA | 15819 | 0.006 | 0.5755 | No |
| 120 | AARS2 | NA | 21456 | -0.001 | 0.4720 | No |
| 121 | MRPL33 | NA | 21919 | -0.002 | 0.4636 | No |
| 122 | MRPL43 | NA | 21967 | -0.002 | 0.4628 | No |
| 123 | RMND1 | NA | 23611 | -0.008 | 0.4330 | No |
| 124 | TFB1M | NA | 23734 | -0.008 | 0.4312 | No |
| 125 | MRPS21 | NA | 23740 | -0.008 | 0.4315 | No |
| 126 | MTG2 | NA | 23913 | -0.009 | 0.4287 | No |
| 127 | MTRF1L | NA | 24047 | -0.009 | 0.4267 | No |
| 128 | MRPS14 | NA | 24335 | -0.010 | 0.4220 | No |
| 129 | SARS2 | NA | 24412 | -0.011 | 0.4211 | No |
| 130 | MRPL42 | NA | 25622 | -0.015 | 0.3996 | No |
| 131 | MRPS27 | NA | 25884 | -0.016 | 0.3956 | No |
| 132 | MRPL34 | NA | 25971 | -0.017 | 0.3949 | No |
| 133 | MRPS25 | NA | 26902 | -0.021 | 0.3788 | No |
| 134 | MRPL45 | NA | 27093 | -0.022 | 0.3764 | No |
| 135 | QRSL1 | NA | 27475 | -0.023 | 0.3705 | No |
| 136 | NOA1 | NA | 28680 | -0.029 | 0.3498 | No |
| 137 | CDK5RAP1 | NA | 29360 | -0.032 | 0.3389 | No |
| 138 | MTIF2 | NA | 30140 | -0.036 | 0.3263 | No |
| 139 | MRPL49 | NA | 30320 | -0.036 | 0.3248 | No |
| 140 | WARS2 | NA | 30762 | -0.038 | 0.3186 | No |
| 141 | MRPL48 | NA | 30834 | -0.039 | 0.3192 | No |
| 142 | OXA1L | NA | 31784 | -0.043 | 0.3038 | No |
| 143 | TEFM | NA | 33256 | -0.051 | 0.2793 | No |
| 144 | TRMT10A | NA | 36347 | -0.067 | 0.2258 | No |
| 145 | RARS2 | NA | 36365 | -0.067 | 0.2288 | No |
| 146 | MRPL53 | NA | 38628 | -0.080 | 0.1912 | No |
| 147 | MRPS6 | NA | 39219 | -0.083 | 0.1844 | No |
| 148 | MRPL55 | NA | 40645 | -0.092 | 0.1627 | No |
| 149 | MTERF1 | NA | 42297 | -0.102 | 0.1374 | No |
| 150 | MTO1 | NA | 42417 | -0.103 | 0.1402 | No |
| 151 | MTERF4 | NA | 43122 | -0.108 | 0.1326 | No |
| 152 | MTIF3 | NA | 44125 | -0.115 | 0.1197 | No |
| 153 | ALKBH1 | NA | 44540 | -0.118 | 0.1179 | No |
| 154 | MRRF | NA | 45415 | -0.124 | 0.1079 | No |
| 155 | FOXO3 | NA | 47286 | -0.139 | 0.0804 | No |
| 156 | PRKAA1 | NA | 47699 | -0.143 | 0.0798 | No |
| 157 | METTL4 | NA | 50065 | -0.166 | 0.0444 | No |
| 158 | NSUN3 | NA | 51875 | -0.189 | 0.0205 | No |
| 159 | TRMT10B | NA | 52393 | -0.198 | 0.0207 | No |
| 160 | MTRF1 | NA | 52482 | -0.200 | 0.0288 | No |
| 161 | MTERF2 | NA | 52615 | -0.203 | 0.0363 | No |