Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_20-29 |
| GeneSet | GOBP_MITOCHONDRIAL_RESPIRATORY_CHAIN_COMPLEX_ASSEMBLY |
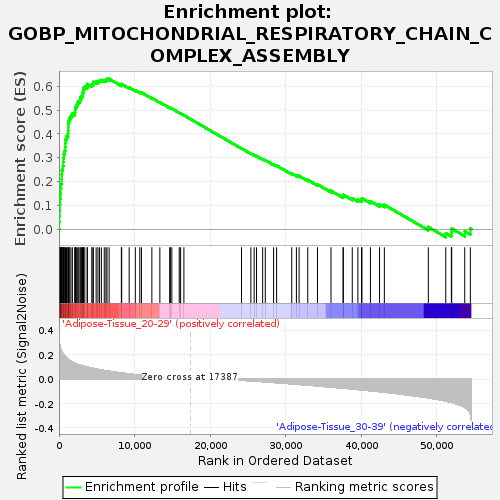
| Enrichment Score (ES) | 0.63089824 |
| Normalized Enrichment Score (NES) | 1.4777436 |
| Nominal p-value | 0.109561756 |
| FDR q-value | 0.51463264 |
| FWER p-Value | 0.966 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SCO1 | NA | 18 | 0.333 | 0.0292 | Yes |
| 2 | NDUFS2 | NA | 42 | 0.301 | 0.0555 | Yes |
| 3 | NDUFAB1 | NA | 80 | 0.286 | 0.0801 | Yes |
| 4 | NDUFAF2 | NA | 101 | 0.279 | 0.1045 | Yes |
| 5 | UQCRFS1 | NA | 139 | 0.267 | 0.1274 | Yes |
| 6 | NDUFAF4 | NA | 193 | 0.254 | 0.1490 | Yes |
| 7 | COA4 | NA | 220 | 0.250 | 0.1706 | Yes |
| 8 | AIFM1 | NA | 270 | 0.242 | 0.1912 | Yes |
| 9 | NDUFA8 | NA | 359 | 0.227 | 0.2097 | Yes |
| 10 | NDUFS5 | NA | 370 | 0.224 | 0.2294 | Yes |
| 11 | NDUFA6 | NA | 385 | 0.223 | 0.2489 | Yes |
| 12 | NDUFA9 | NA | 512 | 0.209 | 0.2650 | Yes |
| 13 | TIMM21 | NA | 562 | 0.203 | 0.2821 | Yes |
| 14 | SCO2 | NA | 584 | 0.201 | 0.2996 | Yes |
| 15 | TMEM126A | NA | 644 | 0.196 | 0.3159 | Yes |
| 16 | NDUFS1 | NA | 761 | 0.188 | 0.3304 | Yes |
| 17 | NDUFAF1 | NA | 832 | 0.183 | 0.3453 | Yes |
| 18 | NDUFS6 | NA | 842 | 0.182 | 0.3613 | Yes |
| 19 | NDUFC2 | NA | 864 | 0.180 | 0.3769 | Yes |
| 20 | FOXRED1 | NA | 988 | 0.173 | 0.3899 | Yes |
| 21 | NDUFV2 | NA | 1184 | 0.162 | 0.4007 | Yes |
| 22 | NDUFB9 | NA | 1210 | 0.160 | 0.4144 | Yes |
| 23 | NDUFB6 | NA | 1229 | 0.159 | 0.4282 | Yes |
| 24 | NDUFA1 | NA | 1232 | 0.159 | 0.4423 | Yes |
| 25 | NDUFAF3 | NA | 1270 | 0.157 | 0.4555 | Yes |
| 26 | SLC25A33 | NA | 1416 | 0.152 | 0.4663 | Yes |
| 27 | NDUFA7 | NA | 1580 | 0.145 | 0.4762 | Yes |
| 28 | TFAM | NA | 1772 | 0.138 | 0.4849 | Yes |
| 29 | NDUFB2 | NA | 2084 | 0.128 | 0.4906 | Yes |
| 30 | CHCHD4 | NA | 2145 | 0.127 | 0.5007 | Yes |
| 31 | NDUFV3 | NA | 2156 | 0.126 | 0.5118 | Yes |
| 32 | NDUFB3 | NA | 2295 | 0.124 | 0.5202 | Yes |
| 33 | TACO1 | NA | 2417 | 0.121 | 0.5287 | Yes |
| 34 | MT-ND6 | NA | 2559 | 0.118 | 0.5366 | Yes |
| 35 | NDUFS3 | NA | 2781 | 0.114 | 0.5426 | Yes |
| 36 | NDUFB8 | NA | 2814 | 0.113 | 0.5521 | Yes |
| 37 | MT-ND5 | NA | 2990 | 0.110 | 0.5586 | Yes |
| 38 | SAMM50 | NA | 3119 | 0.108 | 0.5658 | Yes |
| 39 | NDUFB11 | NA | 3141 | 0.107 | 0.5749 | Yes |
| 40 | NDUFA13 | NA | 3239 | 0.106 | 0.5825 | Yes |
| 41 | NDUFA12 | NA | 3245 | 0.106 | 0.5918 | Yes |
| 42 | MT-ND4 | NA | 3412 | 0.103 | 0.5978 | Yes |
| 43 | NDUFAF8 | NA | 3737 | 0.098 | 0.6006 | Yes |
| 44 | NDUFS8 | NA | 3741 | 0.098 | 0.6092 | Yes |
| 45 | UQCC3 | NA | 4344 | 0.089 | 0.6060 | Yes |
| 46 | NDUFS7 | NA | 4492 | 0.087 | 0.6110 | Yes |
| 47 | COA3 | NA | 4540 | 0.087 | 0.6179 | Yes |
| 48 | NDUFA10 | NA | 4915 | 0.082 | 0.6183 | Yes |
| 49 | UQCC2 | NA | 5154 | 0.079 | 0.6209 | Yes |
| 50 | COX17 | NA | 5365 | 0.077 | 0.6239 | Yes |
| 51 | SDHAF2 | NA | 5610 | 0.074 | 0.6259 | Yes |
| 52 | UQCC1 | NA | 5990 | 0.070 | 0.6252 | Yes |
| 53 | NDUFA3 | NA | 6211 | 0.068 | 0.6271 | Yes |
| 54 | NDUFB10 | NA | 6353 | 0.066 | 0.6304 | Yes |
| 55 | NDUFB7 | NA | 6634 | 0.064 | 0.6309 | Yes |
| 56 | ACAD9 | NA | 8254 | 0.050 | 0.6056 | No |
| 57 | NDUFB1 | NA | 8300 | 0.049 | 0.6092 | No |
| 58 | NDUFA2 | NA | 9287 | 0.042 | 0.5948 | No |
| 59 | ECSIT | NA | 10101 | 0.036 | 0.5831 | No |
| 60 | NDUFS4 | NA | 10698 | 0.032 | 0.5750 | No |
| 61 | COX14 | NA | 10913 | 0.031 | 0.5738 | No |
| 62 | PET117 | NA | 12293 | 0.023 | 0.5505 | No |
| 63 | FASTKD3 | NA | 13352 | 0.017 | 0.5326 | No |
| 64 | PET100 | NA | 14650 | 0.011 | 0.5098 | No |
| 65 | NDUFAF6 | NA | 14741 | 0.011 | 0.5091 | No |
| 66 | SDHAF3 | NA | 14939 | 0.010 | 0.5063 | No |
| 67 | NDUFB4 | NA | 15916 | 0.006 | 0.4889 | No |
| 68 | NDUFB5 | NA | 16097 | 0.005 | 0.4860 | No |
| 69 | COX16 | NA | 16535 | 0.003 | 0.4783 | No |
| 70 | COX20 | NA | 24161 | -0.010 | 0.3392 | No |
| 71 | NDUFV1 | NA | 25399 | -0.014 | 0.3178 | No |
| 72 | NDUFAF5 | NA | 25837 | -0.016 | 0.3112 | No |
| 73 | C12orf73 | NA | 26149 | -0.018 | 0.3071 | No |
| 74 | NDUFC1 | NA | 26958 | -0.021 | 0.2941 | No |
| 75 | TIMMDC1 | NA | 27310 | -0.023 | 0.2897 | No |
| 76 | TTC19 | NA | 28409 | -0.027 | 0.2720 | No |
| 77 | NDUFA11 | NA | 28809 | -0.029 | 0.2672 | No |
| 78 | NDUFAF7 | NA | 30809 | -0.039 | 0.2340 | No |
| 79 | NDUFA5 | NA | 31427 | -0.042 | 0.2264 | No |
| 80 | OXA1L | NA | 31784 | -0.043 | 0.2237 | No |
| 81 | SDHAF1 | NA | 32929 | -0.049 | 0.2070 | No |
| 82 | MT-ND1 | NA | 34219 | -0.056 | 0.1883 | No |
| 83 | COX18 | NA | 36008 | -0.065 | 0.1613 | No |
| 84 | BCS1L | NA | 37605 | -0.074 | 0.1386 | No |
| 85 | COA1 | NA | 37643 | -0.074 | 0.1445 | No |
| 86 | SURF1 | NA | 38823 | -0.081 | 0.1300 | No |
| 87 | SMIM20 | NA | 39568 | -0.085 | 0.1240 | No |
| 88 | SDHAF4 | NA | 40046 | -0.088 | 0.1230 | No |
| 89 | IMMP2L | NA | 40105 | -0.089 | 0.1298 | No |
| 90 | NUBPL | NA | 41226 | -0.095 | 0.1177 | No |
| 91 | LYRM7 | NA | 42432 | -0.103 | 0.1047 | No |
| 92 | MT-ND3 | NA | 43070 | -0.107 | 0.1026 | No |
| 93 | OMA1 | NA | 48892 | -0.154 | 0.0094 | No |
| 94 | COA5 | NA | 51198 | -0.180 | -0.0170 | No |
| 95 | TMEM126B | NA | 51940 | -0.191 | -0.0137 | No |
| 96 | COX19 | NA | 51986 | -0.191 | 0.0024 | No |
| 97 | MT-ND2 | NA | 53712 | -0.232 | -0.0087 | No |
| 98 | TAZ | NA | 54454 | -0.280 | 0.0025 | No |