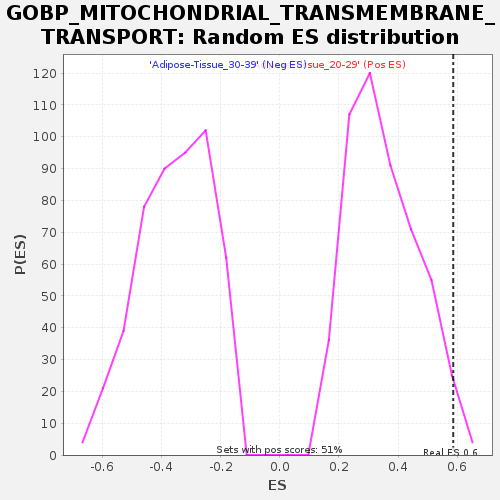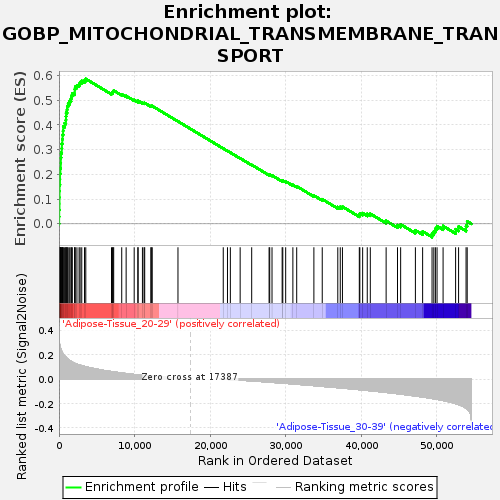
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_20-29 |
| GeneSet | GOBP_MITOCHONDRIAL_TRANSMEMBRANE_TRANSPORT |
| Enrichment Score (ES) | 0.5860528 |
| Normalized Enrichment Score (NES) | 1.6705929 |
| Nominal p-value | 0.021611001 |
| FDR q-value | 0.23006314 |
| FWER p-Value | 0.8 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SLC25A5 | NA | 17 | 0.334 | 0.0287 | Yes |
| 2 | LETM1 | NA | 28 | 0.318 | 0.0562 | Yes |
| 3 | SLC25A22 | NA | 61 | 0.295 | 0.0812 | Yes |
| 4 | TIMM50 | NA | 62 | 0.294 | 0.1068 | Yes |
| 5 | TIMM23 | NA | 67 | 0.292 | 0.1321 | Yes |
| 6 | GRPEL1 | NA | 73 | 0.290 | 0.1571 | Yes |
| 7 | VDAC1 | NA | 122 | 0.272 | 0.1799 | Yes |
| 8 | TOMM40 | NA | 128 | 0.271 | 0.2033 | Yes |
| 9 | TST | NA | 181 | 0.256 | 0.2246 | Yes |
| 10 | AFG3L2 | NA | 228 | 0.249 | 0.2454 | Yes |
| 11 | STOML2 | NA | 234 | 0.247 | 0.2668 | Yes |
| 12 | AIFM1 | NA | 270 | 0.242 | 0.2871 | Yes |
| 13 | MAIP1 | NA | 364 | 0.226 | 0.3051 | Yes |
| 14 | SLC25A15 | NA | 381 | 0.223 | 0.3241 | Yes |
| 15 | TIMM44 | NA | 439 | 0.216 | 0.3419 | Yes |
| 16 | SLC25A51 | NA | 467 | 0.213 | 0.3598 | Yes |
| 17 | HSPD1 | NA | 541 | 0.205 | 0.3763 | Yes |
| 18 | TIMM21 | NA | 562 | 0.203 | 0.3936 | Yes |
| 19 | TIMM17A | NA | 744 | 0.189 | 0.4067 | Yes |
| 20 | SLC25A6 | NA | 903 | 0.178 | 0.4193 | Yes |
| 21 | PNPT1 | NA | 923 | 0.177 | 0.4343 | Yes |
| 22 | SLC25A1 | NA | 944 | 0.176 | 0.4492 | Yes |
| 23 | MPC2 | NA | 1063 | 0.168 | 0.4616 | Yes |
| 24 | DNAJC30 | NA | 1094 | 0.166 | 0.4755 | Yes |
| 25 | ACACB | NA | 1216 | 0.160 | 0.4872 | Yes |
| 26 | SLC25A33 | NA | 1416 | 0.152 | 0.4967 | Yes |
| 27 | PAM16 | NA | 1584 | 0.145 | 0.5062 | Yes |
| 28 | TOMM40L | NA | 1639 | 0.143 | 0.5176 | Yes |
| 29 | SFXN2 | NA | 1762 | 0.139 | 0.5274 | Yes |
| 30 | MPC1 | NA | 2078 | 0.129 | 0.5328 | Yes |
| 31 | ACACA | NA | 2079 | 0.129 | 0.5440 | Yes |
| 32 | CHCHD4 | NA | 2145 | 0.127 | 0.5538 | Yes |
| 33 | TIMM23B | NA | 2374 | 0.122 | 0.5602 | Yes |
| 34 | MID1IP1 | NA | 2660 | 0.116 | 0.5651 | Yes |
| 35 | TOMM70 | NA | 2793 | 0.114 | 0.5725 | Yes |
| 36 | ROMO1 | NA | 3012 | 0.110 | 0.5781 | Yes |
| 37 | UCP2 | NA | 3379 | 0.103 | 0.5803 | Yes |
| 38 | SLC25A20 | NA | 3546 | 0.101 | 0.5861 | Yes |
| 39 | SLC25A32 | NA | 6949 | 0.061 | 0.5289 | No |
| 40 | CPT2 | NA | 7094 | 0.060 | 0.5315 | No |
| 41 | UCP1 | NA | 7142 | 0.059 | 0.5358 | No |
| 42 | SLC25A31 | NA | 7239 | 0.058 | 0.5391 | No |
| 43 | OPA1 | NA | 8304 | 0.049 | 0.5238 | No |
| 44 | SFXN1 | NA | 8888 | 0.045 | 0.5170 | No |
| 45 | MT-ATP6 | NA | 9953 | 0.037 | 0.5008 | No |
| 46 | SFXN5 | NA | 10442 | 0.034 | 0.4948 | No |
| 47 | SLC39A8 | NA | 10467 | 0.034 | 0.4973 | No |
| 48 | MRPL18 | NA | 11044 | 0.030 | 0.4893 | No |
| 49 | TIMM17B | NA | 11255 | 0.029 | 0.4879 | No |
| 50 | SFXN4 | NA | 11345 | 0.028 | 0.4887 | No |
| 51 | TOMM20 | NA | 12157 | 0.023 | 0.4759 | No |
| 52 | SLC25A28 | NA | 12224 | 0.023 | 0.4767 | No |
| 53 | DNAJC15 | NA | 12342 | 0.022 | 0.4764 | No |
| 54 | DNLZ | NA | 15749 | 0.006 | 0.4145 | No |
| 55 | SLC25A37 | NA | 21743 | -0.002 | 0.3047 | No |
| 56 | SLC25A41 | NA | 22303 | -0.003 | 0.2947 | No |
| 57 | MT-ATP8 | NA | 22684 | -0.004 | 0.2881 | No |
| 58 | MRS2 | NA | 23981 | -0.009 | 0.2651 | No |
| 59 | SLC25A52 | NA | 25508 | -0.015 | 0.2384 | No |
| 60 | THRSP | NA | 27811 | -0.025 | 0.1983 | No |
| 61 | PSEN2 | NA | 27894 | -0.025 | 0.1990 | No |
| 62 | SLC25A23 | NA | 28199 | -0.027 | 0.1957 | No |
| 63 | MCUR1 | NA | 29540 | -0.033 | 0.1740 | No |
| 64 | SMDT1 | NA | 29626 | -0.033 | 0.1753 | No |
| 65 | SLC25A4 | NA | 30007 | -0.035 | 0.1714 | No |
| 66 | SLC25A21 | NA | 30960 | -0.039 | 0.1574 | No |
| 67 | MICU2 | NA | 31467 | -0.042 | 0.1517 | No |
| 68 | PRKAG2 | NA | 33741 | -0.053 | 0.1146 | No |
| 69 | SLC25A18 | NA | 34848 | -0.059 | 0.0995 | No |
| 70 | MPC1L | NA | 36906 | -0.070 | 0.0679 | No |
| 71 | SLC25A13 | NA | 37233 | -0.072 | 0.0682 | No |
| 72 | GRPEL2 | NA | 37522 | -0.074 | 0.0693 | No |
| 73 | TOMM7 | NA | 39754 | -0.087 | 0.0359 | No |
| 74 | SLC25A2 | NA | 39825 | -0.087 | 0.0421 | No |
| 75 | MCUB | NA | 40196 | -0.089 | 0.0431 | No |
| 76 | MICU1 | NA | 40807 | -0.093 | 0.0400 | No |
| 77 | UCP3 | NA | 41211 | -0.095 | 0.0409 | No |
| 78 | SLC25A38 | NA | 43305 | -0.109 | 0.0119 | No |
| 79 | DNAJC19 | NA | 44810 | -0.120 | -0.0053 | No |
| 80 | TOMM20L | NA | 45227 | -0.123 | -0.0022 | No |
| 81 | CPT1B | NA | 47179 | -0.138 | -0.0260 | No |
| 82 | PRKAA2 | NA | 48137 | -0.147 | -0.0308 | No |
| 83 | SFXN3 | NA | 49383 | -0.159 | -0.0398 | No |
| 84 | SLC25A36 | NA | 49620 | -0.161 | -0.0302 | No |
| 85 | MCU | NA | 49828 | -0.163 | -0.0198 | No |
| 86 | SLC25A29 | NA | 50046 | -0.166 | -0.0094 | No |
| 87 | PRKAB2 | NA | 50845 | -0.175 | -0.0088 | No |
| 88 | CPT1A | NA | 52503 | -0.200 | -0.0219 | No |
| 89 | SLC25A12 | NA | 52897 | -0.208 | -0.0110 | No |
| 90 | MICU3 | NA | 53886 | -0.238 | -0.0084 | No |
| 91 | SLC8B1 | NA | 54037 | -0.245 | 0.0102 | No |