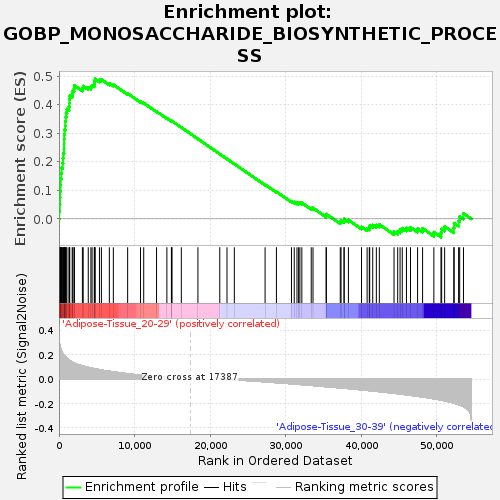
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_20-29 |
| GeneSet | GOBP_MONOSACCHARIDE_BIOSYNTHETIC_PROCESS |
| Enrichment Score (ES) | 0.4907306 |
| Normalized Enrichment Score (NES) | 1.4885179 |
| Nominal p-value | 0.062248997 |
| FDR q-value | 0.5202566 |
| FWER p-Value | 0.965 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | PGM1 | NA | 25 | 0.322 | 0.0280 | Yes |
| 2 | PGD | NA | 103 | 0.277 | 0.0511 | Yes |
| 3 | PCK1 | NA | 127 | 0.271 | 0.0745 | Yes |
| 4 | GPI | NA | 152 | 0.263 | 0.0973 | Yes |
| 5 | ENO1 | NA | 206 | 0.253 | 0.1186 | Yes |
| 6 | GOT1 | NA | 216 | 0.251 | 0.1405 | Yes |
| 7 | MDH2 | NA | 313 | 0.235 | 0.1595 | Yes |
| 8 | TPI1 | NA | 344 | 0.229 | 0.1792 | Yes |
| 9 | PGAM1 | NA | 484 | 0.211 | 0.1952 | Yes |
| 10 | PGK1 | NA | 526 | 0.206 | 0.2127 | Yes |
| 11 | SLC39A14 | NA | 574 | 0.202 | 0.2296 | Yes |
| 12 | RBP4 | NA | 666 | 0.194 | 0.2451 | Yes |
| 13 | SLC25A10 | NA | 673 | 0.194 | 0.2621 | Yes |
| 14 | ADIPOQ | NA | 678 | 0.194 | 0.2791 | Yes |
| 15 | WDR5 | NA | 699 | 0.192 | 0.2957 | Yes |
| 16 | G6PD | NA | 734 | 0.190 | 0.3119 | Yes |
| 17 | GPT | NA | 829 | 0.183 | 0.3263 | Yes |
| 18 | DGAT2 | NA | 838 | 0.182 | 0.3423 | Yes |
| 19 | PC | NA | 898 | 0.179 | 0.3569 | Yes |
| 20 | SLC25A1 | NA | 944 | 0.176 | 0.3716 | Yes |
| 21 | SELENOS | NA | 1046 | 0.169 | 0.3847 | Yes |
| 22 | ALDOA | NA | 1318 | 0.156 | 0.3934 | Yes |
| 23 | PPARA | NA | 1387 | 0.153 | 0.4057 | Yes |
| 24 | SESN2 | NA | 1392 | 0.153 | 0.4191 | Yes |
| 25 | GPD1 | NA | 1419 | 0.151 | 0.4319 | Yes |
| 26 | GAPDH | NA | 1746 | 0.139 | 0.4382 | Yes |
| 27 | SLC25A11 | NA | 1830 | 0.136 | 0.4487 | Yes |
| 28 | TALDO1 | NA | 2003 | 0.131 | 0.4571 | Yes |
| 29 | TKT | NA | 2040 | 0.129 | 0.4679 | Yes |
| 30 | PTPN2 | NA | 3098 | 0.108 | 0.4580 | Yes |
| 31 | ERFE | NA | 3201 | 0.106 | 0.4655 | Yes |
| 32 | PDK2 | NA | 3874 | 0.096 | 0.4617 | Yes |
| 33 | DDB1 | NA | 4226 | 0.091 | 0.4632 | Yes |
| 34 | PFKFB1 | NA | 4377 | 0.089 | 0.4683 | Yes |
| 35 | G6PC3 | NA | 4668 | 0.085 | 0.4705 | Yes |
| 36 | NLN | NA | 4710 | 0.084 | 0.4772 | Yes |
| 37 | CRY1 | NA | 4733 | 0.084 | 0.4842 | Yes |
| 38 | PGP | NA | 4781 | 0.084 | 0.4907 | Yes |
| 39 | SIK1 | NA | 5354 | 0.077 | 0.4870 | No |
| 40 | MDH1 | NA | 5615 | 0.074 | 0.4888 | No |
| 41 | GCK | NA | 6665 | 0.063 | 0.4751 | No |
| 42 | PCK2 | NA | 7193 | 0.059 | 0.4706 | No |
| 43 | SORD | NA | 9087 | 0.043 | 0.4397 | No |
| 44 | C1QTNF12 | NA | 10793 | 0.032 | 0.4112 | No |
| 45 | ALDOC | NA | 11213 | 0.029 | 0.4061 | No |
| 46 | SLC35B4 | NA | 12913 | 0.019 | 0.3766 | No |
| 47 | GAPDHS | NA | 14281 | 0.013 | 0.3526 | No |
| 48 | FBP2 | NA | 14879 | 0.010 | 0.3426 | No |
| 49 | PER2 | NA | 14933 | 0.010 | 0.3424 | No |
| 50 | SDHAF3 | NA | 14939 | 0.010 | 0.3432 | No |
| 51 | MST1 | NA | 16193 | 0.005 | 0.3206 | No |
| 52 | MIR103A1 | NA | 18390 | 0.000 | 0.2803 | No |
| 53 | MIR107 | NA | 21284 | 0.000 | 0.2272 | No |
| 54 | USP7 | NA | 22239 | -0.003 | 0.2100 | No |
| 55 | AKR1A1 | NA | 23209 | -0.006 | 0.1928 | No |
| 56 | ARPP19 | NA | 27281 | -0.022 | 0.1200 | No |
| 57 | PGK2 | NA | 28782 | -0.029 | 0.0951 | No |
| 58 | G6PC2 | NA | 30776 | -0.039 | 0.0619 | No |
| 59 | RANBP2 | NA | 31141 | -0.040 | 0.0588 | No |
| 60 | GOT2 | NA | 31502 | -0.042 | 0.0559 | No |
| 61 | ATF4 | NA | 31719 | -0.043 | 0.0558 | No |
| 62 | SERPINA12 | NA | 31885 | -0.044 | 0.0566 | No |
| 63 | SDS | NA | 32147 | -0.045 | 0.0558 | No |
| 64 | DGKQ | NA | 33390 | -0.051 | 0.0376 | No |
| 65 | SLC37A4 | NA | 33625 | -0.053 | 0.0380 | No |
| 66 | ATF3 | NA | 35361 | -0.062 | 0.0116 | No |
| 67 | FAM3C | NA | 35392 | -0.062 | 0.0165 | No |
| 68 | SLC25A13 | NA | 37233 | -0.072 | -0.0108 | No |
| 69 | INS | NA | 37337 | -0.073 | -0.0063 | No |
| 70 | FAM3A | NA | 37738 | -0.075 | -0.0071 | No |
| 71 | G6PC | NA | 37746 | -0.075 | -0.0006 | No |
| 72 | LEP | NA | 38312 | -0.078 | -0.0041 | No |
| 73 | GCG | NA | 40047 | -0.088 | -0.0281 | No |
| 74 | CHST15 | NA | 40788 | -0.093 | -0.0335 | No |
| 75 | C1QTNF3 | NA | 41076 | -0.094 | -0.0304 | No |
| 76 | MAEA | NA | 41132 | -0.095 | -0.0231 | No |
| 77 | ENO3 | NA | 41531 | -0.097 | -0.0218 | No |
| 78 | SIRT1 | NA | 42010 | -0.100 | -0.0217 | No |
| 79 | GNMT | NA | 42406 | -0.103 | -0.0198 | No |
| 80 | FBP1 | NA | 44350 | -0.116 | -0.0452 | No |
| 81 | ALDOB | NA | 44835 | -0.120 | -0.0435 | No |
| 82 | PPARGC1A | NA | 45150 | -0.122 | -0.0385 | No |
| 83 | AKR1B1 | NA | 45444 | -0.125 | -0.0329 | No |
| 84 | RGN | NA | 45995 | -0.129 | -0.0316 | No |
| 85 | ENO2 | NA | 46529 | -0.133 | -0.0296 | No |
| 86 | PPP4R3B | NA | 47468 | -0.141 | -0.0344 | No |
| 87 | SIRT7 | NA | 48131 | -0.147 | -0.0336 | No |
| 88 | CRTC2 | NA | 49623 | -0.161 | -0.0467 | No |
| 89 | EP300 | NA | 50566 | -0.171 | -0.0489 | No |
| 90 | CLK2 | NA | 50657 | -0.173 | -0.0353 | No |
| 91 | OGT | NA | 51051 | -0.178 | -0.0268 | No |
| 92 | SOGA1 | NA | 52263 | -0.196 | -0.0317 | No |
| 93 | KAT2A | NA | 52308 | -0.196 | -0.0152 | No |
| 94 | SLC25A12 | NA | 52897 | -0.208 | -0.0076 | No |
| 95 | KAT2B | NA | 53043 | -0.212 | 0.0085 | No |
| 96 | LEPR | NA | 53544 | -0.226 | 0.0192 | No |