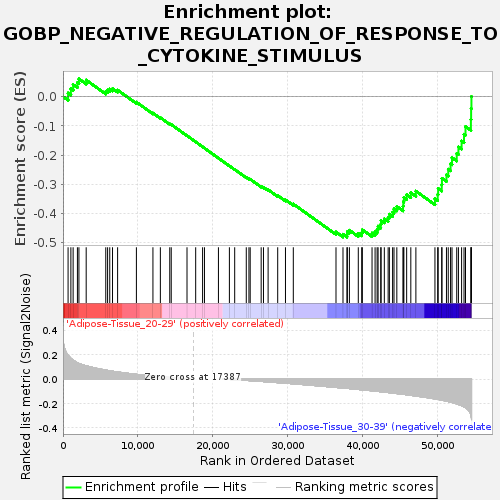
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_30-39 |
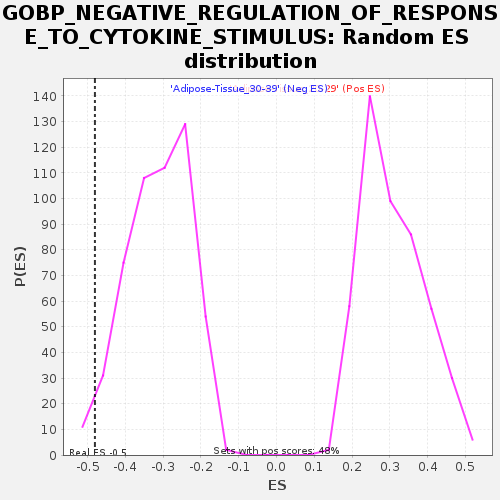
| GeneSet | GOBP_NEGATIVE_REGULATION_OF_RESPONSE_TO_CYTOKINE_STIMULUS |
| Enrichment Score (ES) | -0.48059466 |
| Normalized Enrichment Score (NES) | -1.5410066 |
| Nominal p-value | 0.022988506 |
| FDR q-value | 0.84841496 |
| FWER p-Value | 0.944 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ADIPOQ | NA | 678 | 0.194 | 0.0124 | No |
| 2 | TTLL12 | NA | 1052 | 0.169 | 0.0272 | No |
| 3 | PPARG | NA | 1353 | 0.154 | 0.0414 | No |
| 4 | MIRLET7A1 | NA | 1926 | 0.133 | 0.0480 | No |
| 5 | PXDN | NA | 2127 | 0.127 | 0.0607 | No |
| 6 | PTPN2 | NA | 3098 | 0.108 | 0.0567 | No |
| 7 | YTHDF2 | NA | 5715 | 0.073 | 0.0181 | No |
| 8 | IL6 | NA | 5959 | 0.070 | 0.0226 | No |
| 9 | MUL1 | NA | 6247 | 0.067 | 0.0260 | No |
| 10 | OTUD4 | NA | 6616 | 0.064 | 0.0274 | No |
| 11 | MIR21 | NA | 7297 | 0.058 | 0.0224 | No |
| 12 | MIR27B | NA | 9807 | 0.038 | -0.0187 | No |
| 13 | ARG1 | NA | 12016 | 0.024 | -0.0561 | No |
| 14 | PADI2 | NA | 13009 | 0.019 | -0.0719 | No |
| 15 | PIAS4 | NA | 14265 | 0.013 | -0.0933 | No |
| 16 | MIR135A1 | NA | 14462 | 0.012 | -0.0954 | No |
| 17 | OTOP1 | NA | 16569 | 0.003 | -0.1336 | No |
| 18 | MIR138-1 | NA | 17728 | 0.000 | -0.1548 | No |
| 19 | MIR520C | NA | 18651 | 0.000 | -0.1717 | No |
| 20 | MIR99A | NA | 18901 | 0.000 | -0.1763 | No |
| 21 | MIR26A1 | NA | 20768 | 0.000 | -0.2105 | No |
| 22 | F2RL1 | NA | 22232 | -0.003 | -0.2370 | No |
| 23 | IL1R2 | NA | 22940 | -0.005 | -0.2493 | No |
| 24 | APOA1 | NA | 24488 | -0.011 | -0.2763 | No |
| 25 | MIRLET7C | NA | 24844 | -0.012 | -0.2812 | No |
| 26 | MIR98 | NA | 25012 | -0.013 | -0.2826 | No |
| 27 | MIR125B1 | NA | 26478 | -0.019 | -0.3071 | No |
| 28 | PYDC2 | NA | 26786 | -0.020 | -0.3101 | No |
| 29 | IRAK3 | NA | 27406 | -0.023 | -0.3185 | No |
| 30 | CACTIN | NA | 28687 | -0.029 | -0.3383 | No |
| 31 | MIR152 | NA | 29725 | -0.034 | -0.3530 | No |
| 32 | GSTP1 | NA | 30772 | -0.039 | -0.3672 | No |
| 33 | SH2B3 | NA | 36476 | -0.068 | -0.4631 | No |
| 34 | MIR27A | NA | 37410 | -0.073 | -0.4709 | No |
| 35 | IL36RN | NA | 37942 | -0.076 | -0.4709 | Yes |
| 36 | ROBO1 | NA | 37991 | -0.076 | -0.4620 | Yes |
| 37 | RNF113A | NA | 38267 | -0.078 | -0.4571 | Yes |
| 38 | IL1RN | NA | 39455 | -0.085 | -0.4679 | Yes |
| 39 | GPS2 | NA | 39911 | -0.088 | -0.4651 | Yes |
| 40 | METTL3 | NA | 39997 | -0.088 | -0.4553 | Yes |
| 41 | YTHDF3 | NA | 41292 | -0.096 | -0.4668 | Yes |
| 42 | KLF4 | NA | 41698 | -0.098 | -0.4616 | Yes |
| 43 | SAMHD1 | NA | 41971 | -0.100 | -0.4537 | Yes |
| 44 | PALM3 | NA | 42100 | -0.101 | -0.4431 | Yes |
| 45 | NR1H2 | NA | 42460 | -0.103 | -0.4365 | Yes |
| 46 | NR1H4 | NA | 42498 | -0.103 | -0.4239 | Yes |
| 47 | PYDC1 | NA | 42934 | -0.106 | -0.4182 | Yes |
| 48 | SLIT2 | NA | 43443 | -0.110 | -0.4134 | Yes |
| 49 | MMP12 | NA | 43609 | -0.111 | -0.4022 | Yes |
| 50 | PTPRC | NA | 44039 | -0.114 | -0.3954 | Yes |
| 51 | ADAR | NA | 44225 | -0.115 | -0.3840 | Yes |
| 52 | CCL5 | NA | 44604 | -0.118 | -0.3758 | Yes |
| 53 | CNOT7 | NA | 45431 | -0.124 | -0.3750 | Yes |
| 54 | CAV1 | NA | 45471 | -0.125 | -0.3597 | Yes |
| 55 | MAP2K5 | NA | 45546 | -0.125 | -0.3450 | Yes |
| 56 | NR1H3 | NA | 45917 | -0.128 | -0.3353 | Yes |
| 57 | TRAIP | NA | 46473 | -0.132 | -0.3285 | Yes |
| 58 | NLRP2B | NA | 47150 | -0.138 | -0.3232 | Yes |
| 59 | ECM1 | NA | 49697 | -0.162 | -0.3491 | Yes |
| 60 | MAPK7 | NA | 50064 | -0.166 | -0.3346 | Yes |
| 61 | DCST1 | NA | 50126 | -0.167 | -0.3144 | Yes |
| 62 | NLRC5 | NA | 50605 | -0.172 | -0.3011 | Yes |
| 63 | CARD16 | NA | 50632 | -0.172 | -0.2794 | Yes |
| 64 | CCDC3 | NA | 51235 | -0.180 | -0.2674 | Yes |
| 65 | CARD8 | NA | 51474 | -0.184 | -0.2482 | Yes |
| 66 | PELI3 | NA | 51783 | -0.188 | -0.2297 | Yes |
| 67 | NOL3 | NA | 51977 | -0.191 | -0.2088 | Yes |
| 68 | SLIT3 | NA | 52607 | -0.202 | -0.1943 | Yes |
| 69 | TREX1 | NA | 52834 | -0.207 | -0.1720 | Yes |
| 70 | ZNF675 | NA | 53258 | -0.217 | -0.1519 | Yes |
| 71 | PARP14 | NA | 53581 | -0.227 | -0.1287 | Yes |
| 72 | RFFL | NA | 53780 | -0.234 | -0.1023 | Yes |
| 73 | SIGIRR | NA | 54500 | -0.290 | -0.0783 | Yes |
| 74 | GAS6 | NA | 54531 | -0.299 | -0.0405 | Yes |
| 75 | STAP1 | NA | 54573 | -0.324 | 0.0003 | Yes |