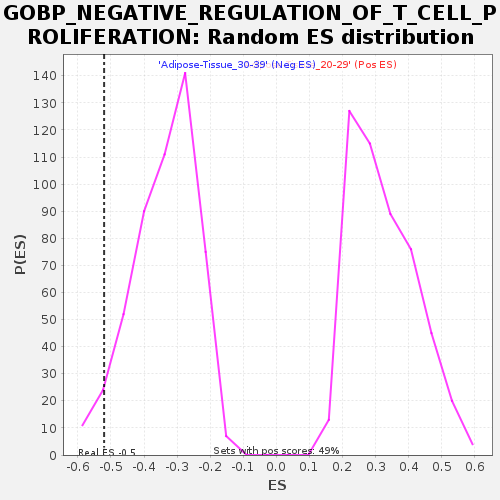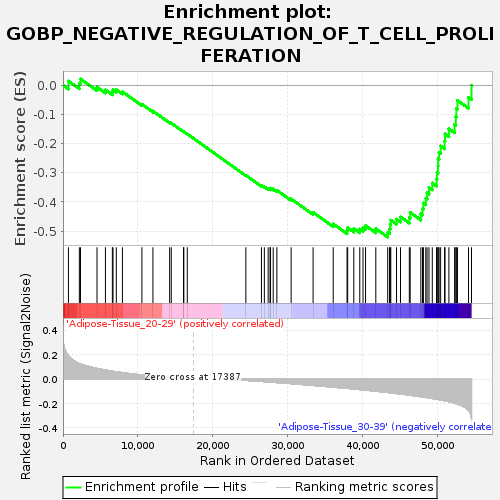
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_30-39 |
| GeneSet | GOBP_NEGATIVE_REGULATION_OF_T_CELL_PROLIFERATION |
| Enrichment Score (ES) | -0.52075654 |
| Normalized Enrichment Score (NES) | -1.5384887 |
| Nominal p-value | 0.04892368 |
| FDR q-value | 0.827027 |
| FWER p-Value | 0.946 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SDC4 | NA | 727 | 0.190 | 0.0147 | No |
| 2 | ARG2 | NA | 2185 | 0.126 | 0.0065 | No |
| 3 | CEBPB | NA | 2335 | 0.123 | 0.0219 | No |
| 4 | TNFRSF21 | NA | 4546 | 0.087 | -0.0059 | No |
| 5 | CLEC4G | NA | 5670 | 0.073 | -0.0158 | No |
| 6 | PRKAR1A | NA | 6595 | 0.064 | -0.0233 | No |
| 7 | ZBTB7B | NA | 6681 | 0.063 | -0.0155 | No |
| 8 | VTCN1 | NA | 7122 | 0.059 | -0.0148 | No |
| 9 | MIR181C | NA | 7938 | 0.052 | -0.0221 | No |
| 10 | FOXJ1 | NA | 10542 | 0.033 | -0.0649 | No |
| 11 | ARG1 | NA | 12016 | 0.024 | -0.0884 | No |
| 12 | GLMN | NA | 14250 | 0.013 | -0.1274 | No |
| 13 | CD274 | NA | 14465 | 0.012 | -0.1296 | No |
| 14 | SFTPD | NA | 16104 | 0.005 | -0.1589 | No |
| 15 | CR1 | NA | 16145 | 0.005 | -0.1589 | No |
| 16 | PRNP | NA | 16604 | 0.003 | -0.1668 | No |
| 17 | IL2RA | NA | 24430 | -0.011 | -0.3088 | No |
| 18 | SCRIB | NA | 26515 | -0.019 | -0.3442 | No |
| 19 | PLA2G2F | NA | 26905 | -0.021 | -0.3483 | No |
| 20 | VSIG4 | NA | 27412 | -0.023 | -0.3542 | No |
| 21 | LILRB2 | NA | 27661 | -0.024 | -0.3552 | No |
| 22 | MAD1L1 | NA | 27731 | -0.024 | -0.3528 | No |
| 23 | IL10 | NA | 28087 | -0.026 | -0.3555 | No |
| 24 | IHH | NA | 28575 | -0.028 | -0.3603 | No |
| 25 | DLG5 | NA | 30482 | -0.037 | -0.3897 | No |
| 26 | SCGB1A1 | NA | 33416 | -0.052 | -0.4359 | No |
| 27 | LGALS7B | NA | 36105 | -0.066 | -0.4755 | No |
| 28 | VSIR | NA | 37952 | -0.076 | -0.4982 | No |
| 29 | FOXP3 | NA | 38039 | -0.077 | -0.4885 | No |
| 30 | LRRC32 | NA | 38856 | -0.081 | -0.4915 | No |
| 31 | HAVCR2 | NA | 39654 | -0.086 | -0.4934 | No |
| 32 | CD86 | NA | 40086 | -0.089 | -0.4883 | No |
| 33 | PLA2G2D | NA | 40425 | -0.091 | -0.4811 | No |
| 34 | CTLA4 | NA | 41796 | -0.099 | -0.4917 | No |
| 35 | SHH | NA | 43384 | -0.109 | -0.5046 | Yes |
| 36 | IL20RB | NA | 43628 | -0.111 | -0.4927 | Yes |
| 37 | LILRB1 | NA | 43720 | -0.112 | -0.4779 | Yes |
| 38 | CBLB | NA | 43798 | -0.112 | -0.4628 | Yes |
| 39 | PAWR | NA | 44565 | -0.118 | -0.4594 | Yes |
| 40 | LILRB4 | NA | 45096 | -0.122 | -0.4512 | Yes |
| 41 | LGALS9 | NA | 46274 | -0.131 | -0.4535 | Yes |
| 42 | LGALS9C | NA | 46387 | -0.132 | -0.4362 | Yes |
| 43 | DLG1 | NA | 47814 | -0.144 | -0.4411 | Yes |
| 44 | PELI1 | NA | 48034 | -0.146 | -0.4235 | Yes |
| 45 | TWSG1 | NA | 48158 | -0.147 | -0.4041 | Yes |
| 46 | BMP4 | NA | 48475 | -0.150 | -0.3878 | Yes |
| 47 | IDO1 | NA | 48642 | -0.151 | -0.3685 | Yes |
| 48 | CD80 | NA | 48902 | -0.154 | -0.3506 | Yes |
| 49 | PTPN6 | NA | 49341 | -0.158 | -0.3353 | Yes |
| 50 | NDFIP1 | NA | 49911 | -0.164 | -0.3216 | Yes |
| 51 | HLA-G | NA | 49970 | -0.165 | -0.2984 | Yes |
| 52 | GNRH1 | NA | 50112 | -0.166 | -0.2765 | Yes |
| 53 | PDCD1LG2 | NA | 50124 | -0.166 | -0.2521 | Yes |
| 54 | CRTAM | NA | 50250 | -0.168 | -0.2297 | Yes |
| 55 | ERBB2 | NA | 50458 | -0.170 | -0.2084 | Yes |
| 56 | SPN | NA | 50985 | -0.177 | -0.1920 | Yes |
| 57 | XCL1 | NA | 51042 | -0.178 | -0.1669 | Yes |
| 58 | LGALS9B | NA | 51562 | -0.185 | -0.1492 | Yes |
| 59 | GPNMB | NA | 52324 | -0.197 | -0.1341 | Yes |
| 60 | CASP3 | NA | 52484 | -0.200 | -0.1076 | Yes |
| 61 | RC3H1 | NA | 52555 | -0.201 | -0.0792 | Yes |
| 62 | HLA-DRB1 | NA | 52708 | -0.204 | -0.0519 | Yes |
| 63 | BTN2A2 | NA | 54185 | -0.253 | -0.0417 | Yes |
| 64 | TNFRSF14 | NA | 54580 | -0.333 | 0.0002 | Yes |