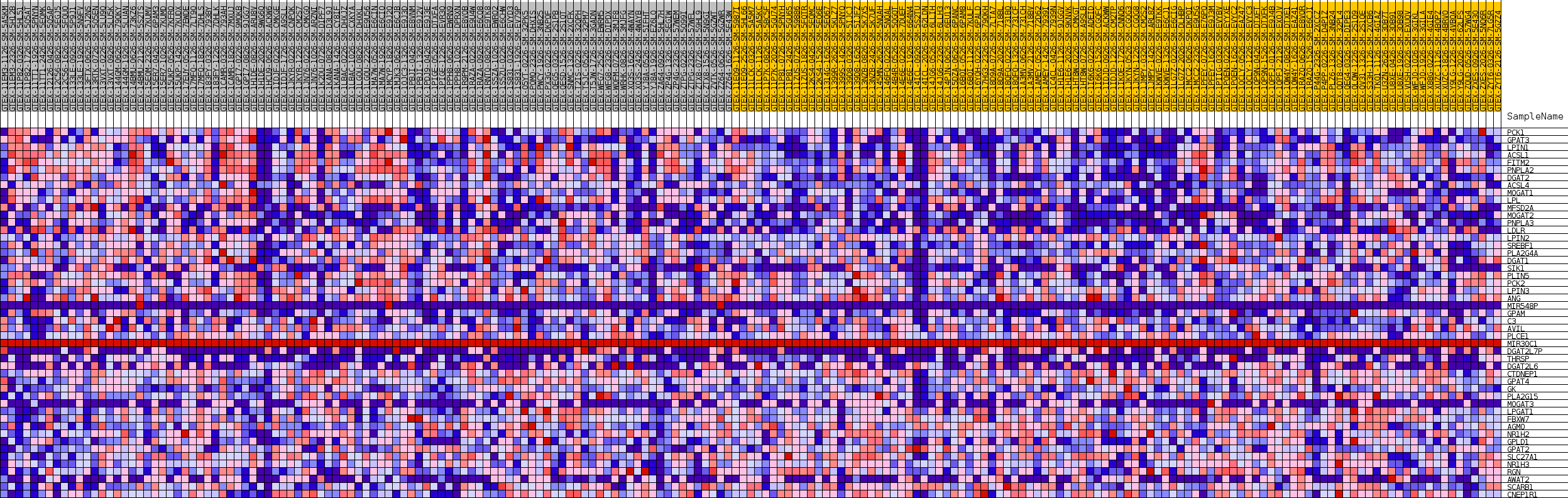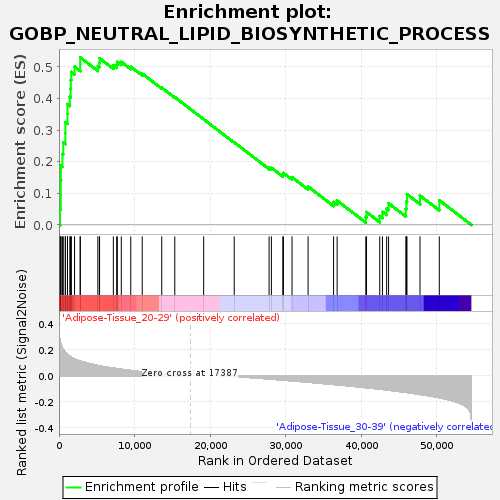
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_20-29 |
| GeneSet | GOBP_NEUTRAL_LIPID_BIOSYNTHETIC_PROCESS |
| Enrichment Score (ES) | 0.53018373 |
| Normalized Enrichment Score (NES) | 1.5523388 |
| Nominal p-value | 0.053719006 |
| FDR q-value | 0.42353103 |
| FWER p-Value | 0.924 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | PCK1 | NA | 127 | 0.271 | 0.0490 | Yes |
| 2 | GPAT3 | NA | 223 | 0.249 | 0.0945 | Yes |
| 3 | LPIN1 | NA | 229 | 0.248 | 0.1415 | Yes |
| 4 | ACSL1 | NA | 252 | 0.244 | 0.1875 | Yes |
| 5 | FITM2 | NA | 453 | 0.214 | 0.2245 | Yes |
| 6 | PNPLA2 | NA | 563 | 0.203 | 0.2610 | Yes |
| 7 | DGAT2 | NA | 838 | 0.182 | 0.2906 | Yes |
| 8 | ACSL4 | NA | 841 | 0.182 | 0.3251 | Yes |
| 9 | MOGAT1 | NA | 1123 | 0.164 | 0.3511 | Yes |
| 10 | LPL | NA | 1126 | 0.164 | 0.3822 | Yes |
| 11 | MFSD2A | NA | 1435 | 0.151 | 0.4051 | Yes |
| 12 | MOGAT2 | NA | 1540 | 0.147 | 0.4310 | Yes |
| 13 | PNPLA3 | NA | 1563 | 0.146 | 0.4583 | Yes |
| 14 | LDLR | NA | 1630 | 0.143 | 0.4843 | Yes |
| 15 | LPIN2 | NA | 2057 | 0.129 | 0.5010 | Yes |
| 16 | SREBF1 | NA | 2809 | 0.113 | 0.5087 | Yes |
| 17 | PLA2G4A | NA | 2812 | 0.113 | 0.5302 | Yes |
| 18 | DGAT1 | NA | 5145 | 0.079 | 0.5025 | No |
| 19 | SIK1 | NA | 5354 | 0.077 | 0.5133 | No |
| 20 | PLIN5 | NA | 5372 | 0.077 | 0.5275 | No |
| 21 | PCK2 | NA | 7193 | 0.059 | 0.5053 | No |
| 22 | LPIN3 | NA | 7636 | 0.055 | 0.5076 | No |
| 23 | ANG | NA | 7721 | 0.054 | 0.5163 | No |
| 24 | MIR548P | NA | 8246 | 0.050 | 0.5162 | No |
| 25 | GPAM | NA | 9499 | 0.040 | 0.5009 | No |
| 26 | C3 | NA | 11019 | 0.030 | 0.4787 | No |
| 27 | AVIL | NA | 13600 | 0.016 | 0.4344 | No |
| 28 | PLCE1 | NA | 15327 | 0.008 | 0.4043 | No |
| 29 | MIR30C1 | NA | 19152 | 0.000 | 0.3342 | No |
| 30 | DGAT2L7P | NA | 23197 | -0.006 | 0.2612 | No |
| 31 | THRSP | NA | 27811 | -0.025 | 0.1813 | No |
| 32 | DGAT2L6 | NA | 28116 | -0.026 | 0.1807 | No |
| 33 | CTDNEP1 | NA | 29627 | -0.033 | 0.1593 | No |
| 34 | GPAT4 | NA | 29709 | -0.034 | 0.1642 | No |
| 35 | GK | NA | 30849 | -0.039 | 0.1507 | No |
| 36 | PLA2G15 | NA | 32974 | -0.049 | 0.1211 | No |
| 37 | MOGAT3 | NA | 36350 | -0.067 | 0.0721 | No |
| 38 | LPGAT1 | NA | 36819 | -0.070 | 0.0767 | No |
| 39 | FBXW7 | NA | 40614 | -0.092 | 0.0246 | No |
| 40 | AGMO | NA | 40711 | -0.092 | 0.0403 | No |
| 41 | NR1H2 | NA | 42460 | -0.103 | 0.0279 | No |
| 42 | GPLD1 | NA | 42817 | -0.106 | 0.0414 | No |
| 43 | GPAT2 | NA | 43374 | -0.109 | 0.0519 | No |
| 44 | SLC27A1 | NA | 43630 | -0.111 | 0.0683 | No |
| 45 | NR1H3 | NA | 45917 | -0.128 | 0.0507 | No |
| 46 | RGN | NA | 45995 | -0.129 | 0.0737 | No |
| 47 | AWAT2 | NA | 46062 | -0.129 | 0.0970 | No |
| 48 | SCARB1 | NA | 47791 | -0.144 | 0.0926 | No |
| 49 | CNEP1R1 | NA | 50345 | -0.169 | 0.0778 | No |