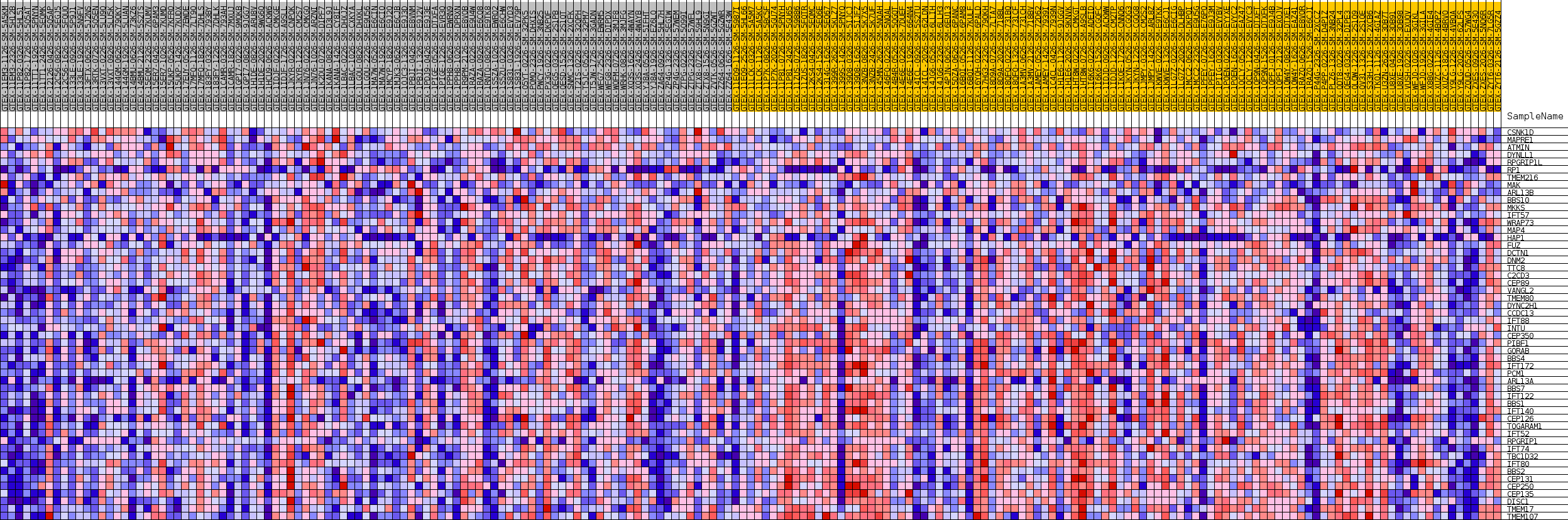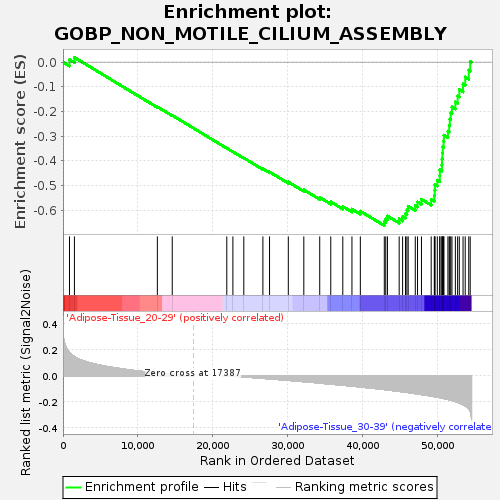
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_30-39 |
| GeneSet | GOBP_NON_MOTILE_CILIUM_ASSEMBLY |
| Enrichment Score (ES) | -0.6631697 |
| Normalized Enrichment Score (NES) | -1.605992 |
| Nominal p-value | 0.015904572 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.885 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | CSNK1D | NA | 868 | 0.180 | 0.0100 | No |
| 2 | MAPRE1 | NA | 1526 | 0.147 | 0.0191 | No |
| 3 | ATMIN | NA | 12607 | 0.021 | -0.1810 | No |
| 4 | DYNLL1 | NA | 14596 | 0.011 | -0.2159 | No |
| 5 | RPGRIP1L | NA | 21885 | -0.002 | -0.3492 | No |
| 6 | RP1 | NA | 22699 | -0.005 | -0.3635 | No |
| 7 | TMEM216 | NA | 24158 | -0.010 | -0.3888 | No |
| 8 | MAK | NA | 26708 | -0.020 | -0.4327 | No |
| 9 | ARL13B | NA | 27598 | -0.024 | -0.4456 | No |
| 10 | BBS10 | NA | 30104 | -0.035 | -0.4864 | No |
| 11 | MKKS | NA | 32176 | -0.045 | -0.5179 | No |
| 12 | IFT57 | NA | 34302 | -0.056 | -0.5487 | No |
| 13 | WRAP73 | NA | 35777 | -0.064 | -0.5665 | No |
| 14 | MAP4 | NA | 37385 | -0.073 | -0.5855 | No |
| 15 | HAP1 | NA | 38607 | -0.080 | -0.5964 | No |
| 16 | FUZ | NA | 39728 | -0.086 | -0.6045 | No |
| 17 | DCTN1 | NA | 42930 | -0.106 | -0.6479 | Yes |
| 18 | DNM2 | NA | 43125 | -0.108 | -0.6359 | Yes |
| 19 | TTC8 | NA | 43349 | -0.109 | -0.6243 | Yes |
| 20 | C2CD3 | NA | 44918 | -0.121 | -0.6357 | Yes |
| 21 | CEP89 | NA | 45376 | -0.124 | -0.6263 | Yes |
| 22 | VANGL2 | NA | 45759 | -0.127 | -0.6150 | Yes |
| 23 | TMEM80 | NA | 45948 | -0.128 | -0.6000 | Yes |
| 24 | DYNC2H1 | NA | 46122 | -0.130 | -0.5846 | Yes |
| 25 | CCDC13 | NA | 47069 | -0.138 | -0.5821 | Yes |
| 26 | IFT88 | NA | 47372 | -0.140 | -0.5675 | Yes |
| 27 | INTU | NA | 47895 | -0.145 | -0.5562 | Yes |
| 28 | CEP350 | NA | 49195 | -0.157 | -0.5575 | Yes |
| 29 | PIBF1 | NA | 49610 | -0.161 | -0.5420 | Yes |
| 30 | GORAB | NA | 49668 | -0.162 | -0.5198 | Yes |
| 31 | BBS4 | NA | 49701 | -0.162 | -0.4971 | Yes |
| 32 | IFT172 | NA | 50014 | -0.165 | -0.4791 | Yes |
| 33 | PCM1 | NA | 50347 | -0.169 | -0.4609 | Yes |
| 34 | ARL13A | NA | 50354 | -0.169 | -0.4367 | Yes |
| 35 | BBS7 | NA | 50624 | -0.172 | -0.4168 | Yes |
| 36 | IFT122 | NA | 50648 | -0.173 | -0.3924 | Yes |
| 37 | BBS1 | NA | 50700 | -0.173 | -0.3685 | Yes |
| 38 | IFT140 | NA | 50735 | -0.174 | -0.3441 | Yes |
| 39 | CEP126 | NA | 50858 | -0.175 | -0.3212 | Yes |
| 40 | TOGARAM1 | NA | 50889 | -0.176 | -0.2965 | Yes |
| 41 | IFT52 | NA | 51431 | -0.183 | -0.2801 | Yes |
| 42 | RPGRIP1 | NA | 51600 | -0.185 | -0.2565 | Yes |
| 43 | IFT74 | NA | 51683 | -0.186 | -0.2312 | Yes |
| 44 | TBC1D32 | NA | 51810 | -0.188 | -0.2064 | Yes |
| 45 | IFT80 | NA | 51971 | -0.191 | -0.1819 | Yes |
| 46 | BBS2 | NA | 52424 | -0.199 | -0.1616 | Yes |
| 47 | CEP131 | NA | 52727 | -0.205 | -0.1377 | Yes |
| 48 | CEP250 | NA | 52952 | -0.210 | -0.1116 | Yes |
| 49 | CEP135 | NA | 53450 | -0.222 | -0.0887 | Yes |
| 50 | DISC1 | NA | 53741 | -0.233 | -0.0606 | Yes |
| 51 | TMEM17 | NA | 54213 | -0.255 | -0.0326 | Yes |
| 52 | TMEM107 | NA | 54408 | -0.275 | 0.0034 | Yes |