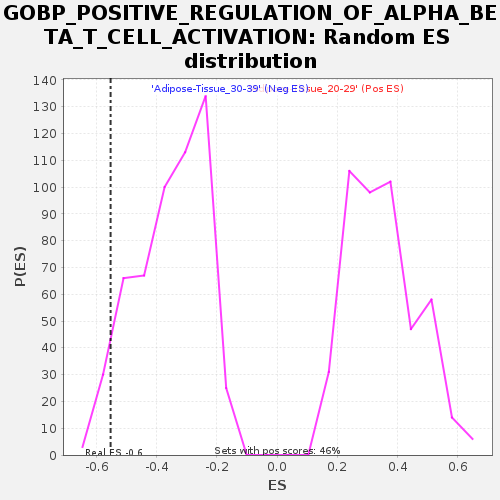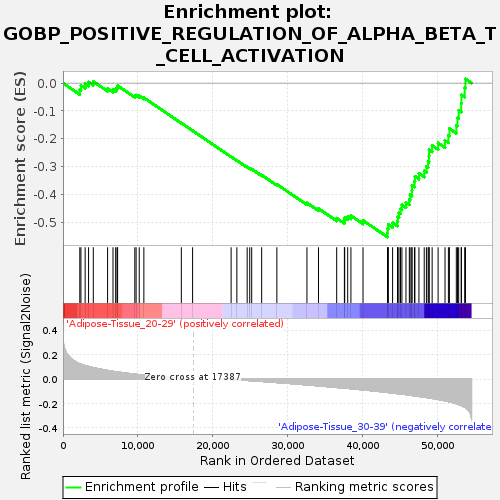
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_30-39 |
| GeneSet | GOBP_POSITIVE_REGULATION_OF_ALPHA_BETA_T_CELL_ACTIVATION |
| Enrichment Score (ES) | -0.553295 |
| Normalized Enrichment Score (NES) | -1.5648206 |
| Nominal p-value | 0.050185874 |
| FDR q-value | 0.92721695 |
| FWER p-Value | 0.927 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | AP3D1 | NA | 2245 | 0.125 | -0.0232 | No |
| 2 | PNP | NA | 2395 | 0.121 | -0.0084 | No |
| 3 | RUNX1 | NA | 2958 | 0.110 | -0.0028 | No |
| 4 | SOCS1 | NA | 3414 | 0.103 | 0.0037 | No |
| 5 | CD81 | NA | 4048 | 0.093 | 0.0055 | No |
| 6 | IL12A | NA | 5958 | 0.070 | -0.0193 | No |
| 7 | ZBTB7B | NA | 6681 | 0.063 | -0.0235 | No |
| 8 | CD55 | NA | 7037 | 0.060 | -0.0213 | No |
| 9 | RARA | NA | 7217 | 0.059 | -0.0161 | No |
| 10 | MIR21 | NA | 7297 | 0.058 | -0.0092 | No |
| 11 | HSPH1 | NA | 9596 | 0.040 | -0.0456 | No |
| 12 | ANXA1 | NA | 9769 | 0.039 | -0.0432 | No |
| 13 | NFKBIZ | NA | 10185 | 0.036 | -0.0457 | No |
| 14 | IL23A | NA | 10803 | 0.032 | -0.0524 | No |
| 15 | GLI3 | NA | 15809 | 0.006 | -0.1433 | No |
| 16 | IL4R | NA | 17317 | 0.000 | -0.1709 | No |
| 17 | IL12B | NA | 22462 | -0.004 | -0.2647 | No |
| 18 | CBFB | NA | 23231 | -0.006 | -0.2779 | No |
| 19 | PRKCZ | NA | 24607 | -0.011 | -0.3015 | No |
| 20 | AP3B1 | NA | 24951 | -0.012 | -0.3060 | No |
| 21 | NFKBID | NA | 25213 | -0.013 | -0.3088 | No |
| 22 | CCR2 | NA | 26543 | -0.019 | -0.3304 | No |
| 23 | IHH | NA | 28575 | -0.028 | -0.3636 | No |
| 24 | ITPKB | NA | 32593 | -0.047 | -0.4304 | No |
| 25 | IL23R | NA | 34139 | -0.055 | -0.4508 | No |
| 26 | RIPK2 | NA | 36568 | -0.069 | -0.4854 | No |
| 27 | CCL19 | NA | 37597 | -0.074 | -0.4936 | No |
| 28 | HLX | NA | 37639 | -0.074 | -0.4836 | No |
| 29 | FOXP3 | NA | 38039 | -0.077 | -0.4799 | No |
| 30 | NLRP3 | NA | 38469 | -0.079 | -0.4764 | No |
| 31 | CD86 | NA | 40086 | -0.089 | -0.4933 | No |
| 32 | SOCS5 | NA | 43359 | -0.109 | -0.5375 | Yes |
| 33 | SHH | NA | 43384 | -0.109 | -0.5222 | Yes |
| 34 | CD83 | NA | 43468 | -0.110 | -0.5079 | Yes |
| 35 | PTPRC | NA | 44039 | -0.114 | -0.5019 | Yes |
| 36 | SASH3 | NA | 44692 | -0.119 | -0.4967 | Yes |
| 37 | TNFSF4 | NA | 44738 | -0.119 | -0.4803 | Yes |
| 38 | TGFBR2 | NA | 44890 | -0.120 | -0.4657 | Yes |
| 39 | LILRB4 | NA | 45096 | -0.122 | -0.4519 | Yes |
| 40 | ZBTB16 | NA | 45285 | -0.123 | -0.4376 | Yes |
| 41 | CD28 | NA | 45825 | -0.127 | -0.4291 | Yes |
| 42 | LGALS9 | NA | 46274 | -0.131 | -0.4185 | Yes |
| 43 | CD3E | NA | 46377 | -0.132 | -0.4013 | Yes |
| 44 | IL12RB1 | NA | 46615 | -0.134 | -0.3864 | Yes |
| 45 | ADA | NA | 46625 | -0.134 | -0.3672 | Yes |
| 46 | NKAP | NA | 46960 | -0.137 | -0.3536 | Yes |
| 47 | IL18 | NA | 47019 | -0.137 | -0.3349 | Yes |
| 48 | RASAL3 | NA | 47558 | -0.142 | -0.3243 | Yes |
| 49 | EBI3 | NA | 48264 | -0.148 | -0.3159 | Yes |
| 50 | PTPN22 | NA | 48559 | -0.151 | -0.2996 | Yes |
| 51 | NCKAP1L | NA | 48786 | -0.153 | -0.2817 | Yes |
| 52 | CD80 | NA | 48902 | -0.154 | -0.2616 | Yes |
| 53 | SYK | NA | 48916 | -0.154 | -0.2397 | Yes |
| 54 | HLA-A | NA | 49322 | -0.158 | -0.2243 | Yes |
| 55 | MYB | NA | 50119 | -0.166 | -0.2149 | Yes |
| 56 | XCL1 | NA | 51042 | -0.178 | -0.2062 | Yes |
| 57 | ZAP70 | NA | 51495 | -0.184 | -0.1879 | Yes |
| 58 | RUNX3 | NA | 51629 | -0.186 | -0.1636 | Yes |
| 59 | IFNG | NA | 52552 | -0.201 | -0.1515 | Yes |
| 60 | HLA-DRB1 | NA | 52708 | -0.204 | -0.1249 | Yes |
| 61 | HLA-DRA | NA | 52868 | -0.208 | -0.0978 | Yes |
| 62 | MALT1 | NA | 53209 | -0.216 | -0.0729 | Yes |
| 63 | CD160 | NA | 53232 | -0.217 | -0.0421 | Yes |
| 64 | PRKCQ | NA | 53674 | -0.231 | -0.0169 | Yes |
| 65 | HLA-E | NA | 53772 | -0.234 | 0.0150 | Yes |