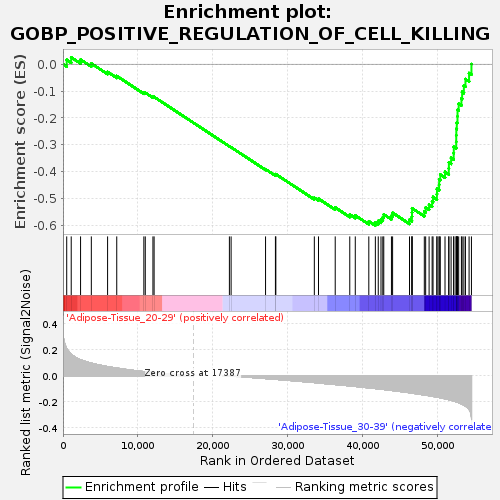
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_30-39 |
| GeneSet | GOBP_POSITIVE_REGULATION_OF_CELL_KILLING |
| Enrichment Score (ES) | -0.60184133 |
| Normalized Enrichment Score (NES) | -1.6799328 |
| Nominal p-value | 0.0151802655 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.758 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | FADD | NA | 496 | 0.210 | 0.0163 | No |
| 2 | AZGP1 | NA | 1098 | 0.165 | 0.0254 | No |
| 3 | PVR | NA | 2347 | 0.122 | 0.0173 | No |
| 4 | SLC22A13 | NA | 3784 | 0.097 | 0.0027 | No |
| 5 | IL12A | NA | 5958 | 0.070 | -0.0286 | No |
| 6 | NECTIN2 | NA | 7181 | 0.059 | -0.0439 | No |
| 7 | IL23A | NA | 10803 | 0.032 | -0.1064 | No |
| 8 | LAG3 | NA | 10993 | 0.030 | -0.1062 | No |
| 9 | ARG1 | NA | 12016 | 0.024 | -0.1220 | No |
| 10 | CADM1 | NA | 12172 | 0.023 | -0.1220 | No |
| 11 | F2RL1 | NA | 22232 | -0.003 | -0.3061 | No |
| 12 | IL12B | NA | 22462 | -0.004 | -0.3099 | No |
| 13 | RAET1E | NA | 27064 | -0.021 | -0.3916 | No |
| 14 | IL18RAP | NA | 28382 | -0.027 | -0.4125 | No |
| 15 | CD1D | NA | 28438 | -0.028 | -0.4101 | No |
| 16 | VAV1 | NA | 33581 | -0.052 | -0.4980 | No |
| 17 | IL23R | NA | 34139 | -0.055 | -0.5015 | No |
| 18 | HLA-H | NA | 36372 | -0.067 | -0.5343 | No |
| 19 | IL21 | NA | 38317 | -0.078 | -0.5605 | No |
| 20 | CD5L | NA | 39052 | -0.082 | -0.5640 | No |
| 21 | STAT5B | NA | 40860 | -0.093 | -0.5858 | No |
| 22 | ITGAM | NA | 41735 | -0.099 | -0.5899 | Yes |
| 23 | NOS2 | NA | 42122 | -0.101 | -0.5847 | Yes |
| 24 | AP1G1 | NA | 42488 | -0.103 | -0.5788 | Yes |
| 25 | NCR3 | NA | 42731 | -0.105 | -0.5706 | Yes |
| 26 | CD1A | NA | 42859 | -0.106 | -0.5600 | Yes |
| 27 | FCER2 | NA | 43885 | -0.113 | -0.5651 | Yes |
| 28 | PTPRC | NA | 44039 | -0.114 | -0.5541 | Yes |
| 29 | TYROBP | NA | 46299 | -0.131 | -0.5796 | Yes |
| 30 | IL12RB1 | NA | 46615 | -0.134 | -0.5692 | Yes |
| 31 | RASGRP1 | NA | 46632 | -0.134 | -0.5533 | Yes |
| 32 | KLRC2 | NA | 46646 | -0.134 | -0.5373 | Yes |
| 33 | STX7 | NA | 48276 | -0.148 | -0.5492 | Yes |
| 34 | HLA-B | NA | 48457 | -0.150 | -0.5343 | Yes |
| 35 | SYK | NA | 48916 | -0.154 | -0.5240 | Yes |
| 36 | HLA-A | NA | 49322 | -0.158 | -0.5123 | Yes |
| 37 | BAD | NA | 49440 | -0.159 | -0.4951 | Yes |
| 38 | CLEC7A | NA | 49956 | -0.165 | -0.4846 | Yes |
| 39 | HLA-G | NA | 49970 | -0.165 | -0.4649 | Yes |
| 40 | CRTAM | NA | 50250 | -0.168 | -0.4497 | Yes |
| 41 | SLAMF6 | NA | 50263 | -0.168 | -0.4295 | Yes |
| 42 | LAMP1 | NA | 50411 | -0.170 | -0.4117 | Yes |
| 43 | XCL1 | NA | 51042 | -0.178 | -0.4017 | Yes |
| 44 | PRF1 | NA | 51544 | -0.185 | -0.3885 | Yes |
| 45 | BCL2L11 | NA | 51558 | -0.185 | -0.3663 | Yes |
| 46 | CD1C | NA | 51855 | -0.189 | -0.3489 | Yes |
| 47 | KLRD1 | NA | 52199 | -0.195 | -0.3315 | Yes |
| 48 | CD226 | NA | 52216 | -0.195 | -0.3082 | Yes |
| 49 | SH2D1A | NA | 52536 | -0.201 | -0.2897 | Yes |
| 50 | B2M | NA | 52543 | -0.201 | -0.2654 | Yes |
| 51 | IFNG | NA | 52552 | -0.201 | -0.2412 | Yes |
| 52 | CD1B | NA | 52616 | -0.203 | -0.2178 | Yes |
| 53 | HLA-DRB1 | NA | 52708 | -0.204 | -0.1947 | Yes |
| 54 | HLA-F | NA | 52715 | -0.204 | -0.1701 | Yes |
| 55 | HLA-DRA | NA | 52868 | -0.208 | -0.1477 | Yes |
| 56 | CD160 | NA | 53232 | -0.217 | -0.1281 | Yes |
| 57 | KLRK1 | NA | 53328 | -0.219 | -0.1032 | Yes |
| 58 | MR1 | NA | 53563 | -0.226 | -0.0801 | Yes |
| 59 | HLA-E | NA | 53772 | -0.234 | -0.0556 | Yes |
| 60 | CD1E | NA | 54258 | -0.258 | -0.0332 | Yes |
| 61 | STAP1 | NA | 54573 | -0.324 | 0.0003 | Yes |

