Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
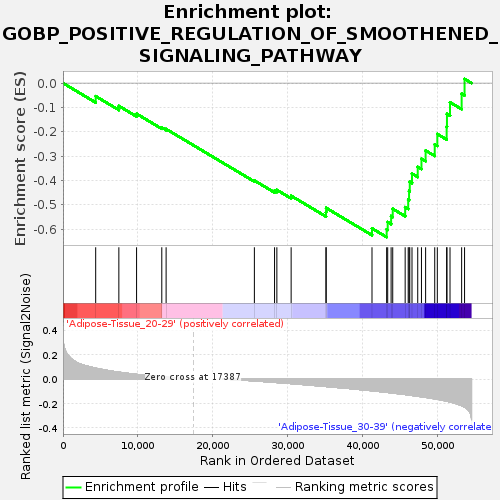
| Upregulated in class | Adipose-Tissue_30-39 |
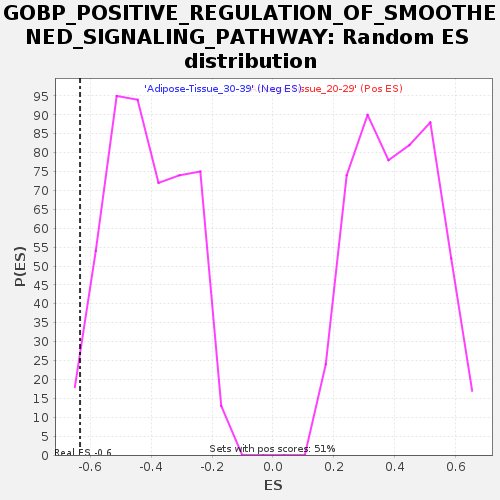
| GeneSet | GOBP_POSITIVE_REGULATION_OF_SMOOTHENED_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.6330084 |
| Normalized Enrichment Score (NES) | -1.5395435 |
| Nominal p-value | 0.018181818 |
| FDR q-value | 0.83844393 |
| FWER p-Value | 0.944 |

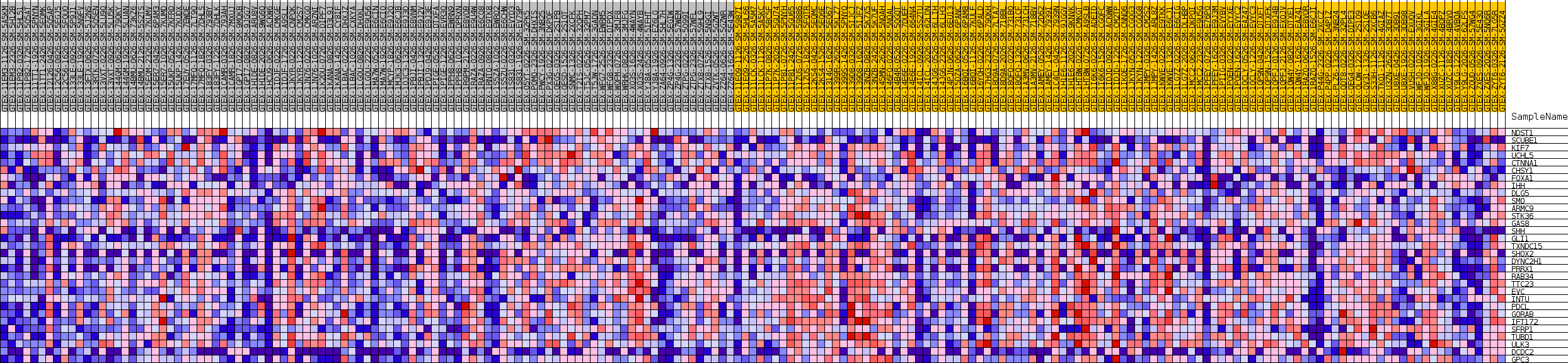
| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | NDST1 | NA | 4367 | 0.089 | -0.0539 | No |
| 2 | SCUBE1 | NA | 7463 | 0.056 | -0.0940 | No |
| 3 | KIF7 | NA | 9831 | 0.038 | -0.1261 | No |
| 4 | UCHL5 | NA | 13200 | 0.018 | -0.1826 | No |
| 5 | CTNNA1 | NA | 13777 | 0.015 | -0.1888 | No |
| 6 | CHSY1 | NA | 25570 | -0.015 | -0.4005 | No |
| 7 | FOXA1 | NA | 28266 | -0.027 | -0.4420 | No |
| 8 | IHH | NA | 28575 | -0.028 | -0.4393 | No |
| 9 | DLG5 | NA | 30482 | -0.037 | -0.4633 | No |
| 10 | SMO | NA | 35137 | -0.061 | -0.5306 | No |
| 11 | ARMC9 | NA | 35160 | -0.061 | -0.5131 | No |
| 12 | STK36 | NA | 41286 | -0.096 | -0.5971 | No |
| 13 | GAS8 | NA | 43244 | -0.109 | -0.6010 | Yes |
| 14 | SHH | NA | 43384 | -0.109 | -0.5714 | Yes |
| 15 | GLI1 | NA | 43840 | -0.113 | -0.5465 | Yes |
| 16 | TXNDC15 | NA | 44056 | -0.114 | -0.5168 | Yes |
| 17 | SHOX2 | NA | 45717 | -0.127 | -0.5100 | Yes |
| 18 | DYNC2H1 | NA | 46122 | -0.130 | -0.4792 | Yes |
| 19 | PRRX1 | NA | 46240 | -0.131 | -0.4429 | Yes |
| 20 | RAB34 | NA | 46333 | -0.131 | -0.4059 | Yes |
| 21 | TTC23 | NA | 46631 | -0.134 | -0.3719 | Yes |
| 22 | EVC | NA | 47397 | -0.140 | -0.3445 | Yes |
| 23 | INTU | NA | 47895 | -0.145 | -0.3110 | Yes |
| 24 | PDCL | NA | 48451 | -0.150 | -0.2771 | Yes |
| 25 | GORAB | NA | 49668 | -0.162 | -0.2518 | Yes |
| 26 | IFT172 | NA | 50014 | -0.165 | -0.2094 | Yes |
| 27 | SFRP1 | NA | 51261 | -0.180 | -0.1791 | Yes |
| 28 | TUBD1 | NA | 51302 | -0.181 | -0.1265 | Yes |
| 29 | ULK3 | NA | 51710 | -0.187 | -0.0789 | Yes |
| 30 | DCDC2 | NA | 53264 | -0.217 | -0.0434 | Yes |
| 31 | GPC3 | NA | 53654 | -0.230 | 0.0172 | Yes |