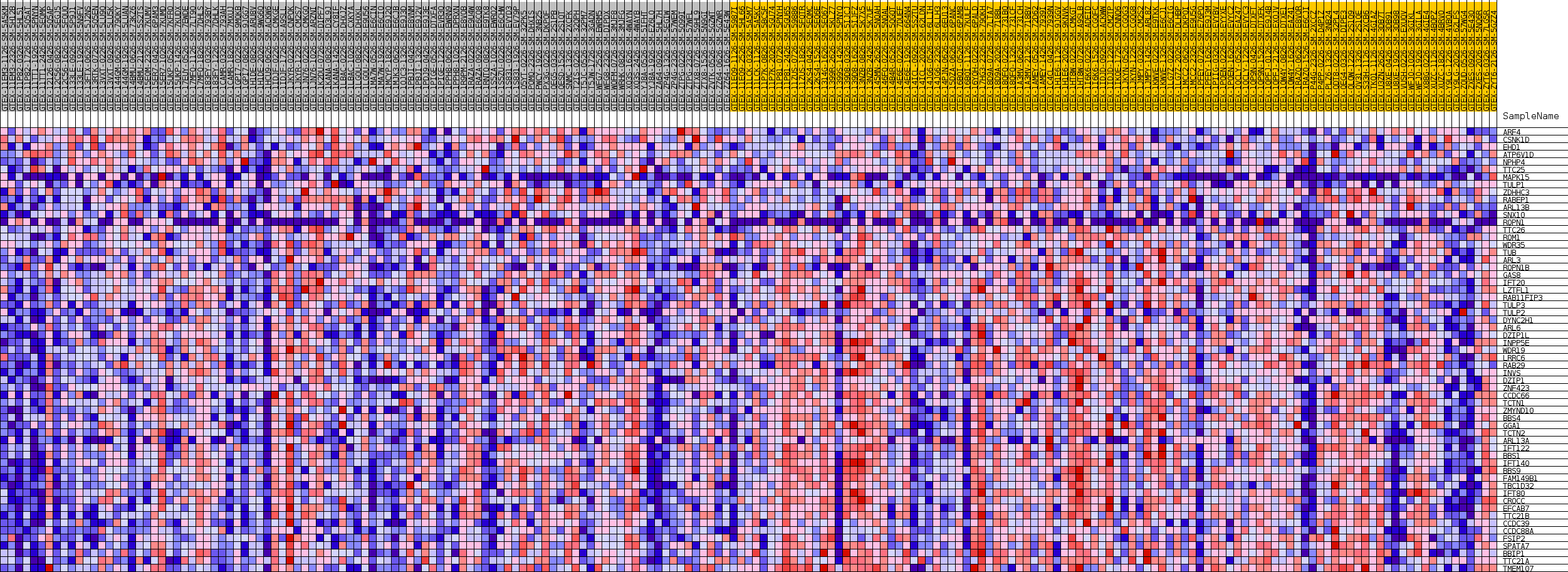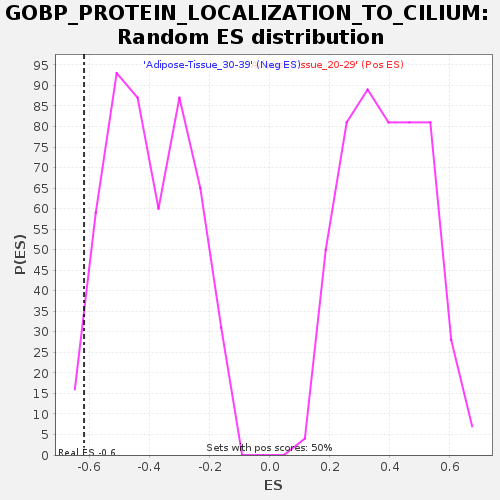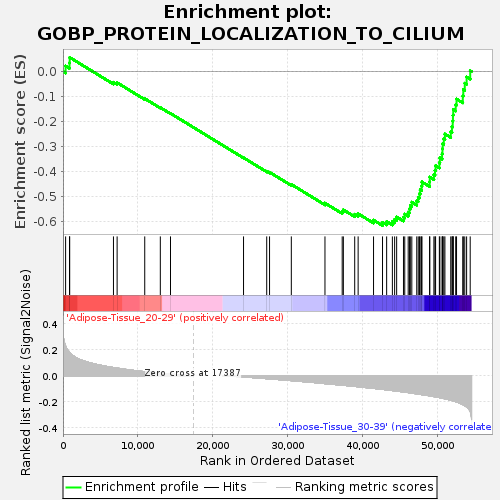
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_30-39 |
| GeneSet | GOBP_PROTEIN_LOCALIZATION_TO_CILIUM |
| Enrichment Score (ES) | -0.6176464 |
| Normalized Enrichment Score (NES) | -1.5515096 |
| Nominal p-value | 0.024096385 |
| FDR q-value | 0.86600393 |
| FWER p-Value | 0.936 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ARF4 | NA | 349 | 0.228 | 0.0219 | No |
| 2 | CSNK1D | NA | 868 | 0.180 | 0.0346 | No |
| 3 | EHD1 | NA | 895 | 0.179 | 0.0563 | No |
| 4 | ATP6V1D | NA | 6742 | 0.063 | -0.0432 | No |
| 5 | NPHP4 | NA | 7231 | 0.058 | -0.0449 | No |
| 6 | TTC25 | NA | 10921 | 0.031 | -0.1087 | No |
| 7 | MAPK15 | NA | 13003 | 0.019 | -0.1446 | No |
| 8 | TULP1 | NA | 14363 | 0.012 | -0.1680 | No |
| 9 | ZDHHC3 | NA | 24122 | -0.009 | -0.3457 | No |
| 10 | RABEP1 | NA | 27219 | -0.022 | -0.3998 | No |
| 11 | ARL13B | NA | 27598 | -0.024 | -0.4037 | No |
| 12 | SNX10 | NA | 30506 | -0.037 | -0.4524 | No |
| 13 | ROPN1 | NA | 35002 | -0.060 | -0.5274 | No |
| 14 | TTC26 | NA | 37304 | -0.073 | -0.5606 | No |
| 15 | ROM1 | NA | 37460 | -0.073 | -0.5544 | No |
| 16 | WDR35 | NA | 38976 | -0.082 | -0.5720 | No |
| 17 | TUB | NA | 39434 | -0.085 | -0.5700 | No |
| 18 | ARL3 | NA | 41492 | -0.097 | -0.5957 | No |
| 19 | ROPN1B | NA | 42692 | -0.105 | -0.6047 | Yes |
| 20 | GAS8 | NA | 43244 | -0.109 | -0.6014 | Yes |
| 21 | IFT20 | NA | 44009 | -0.114 | -0.6013 | Yes |
| 22 | LZTFL1 | NA | 44311 | -0.116 | -0.5925 | Yes |
| 23 | RAB11FIP3 | NA | 44581 | -0.118 | -0.5828 | Yes |
| 24 | TULP3 | NA | 45492 | -0.125 | -0.5840 | Yes |
| 25 | TULP2 | NA | 45663 | -0.126 | -0.5715 | Yes |
| 26 | DYNC2H1 | NA | 46122 | -0.130 | -0.5639 | Yes |
| 27 | ARL6 | NA | 46290 | -0.131 | -0.5507 | Yes |
| 28 | DZIP1L | NA | 46406 | -0.132 | -0.5365 | Yes |
| 29 | INPP5E | NA | 46606 | -0.134 | -0.5236 | Yes |
| 30 | WDR19 | NA | 47267 | -0.139 | -0.5185 | Yes |
| 31 | LRRC6 | NA | 47464 | -0.141 | -0.5047 | Yes |
| 32 | RAB29 | NA | 47627 | -0.142 | -0.4900 | Yes |
| 33 | INVS | NA | 47720 | -0.143 | -0.4740 | Yes |
| 34 | DZIP1 | NA | 47909 | -0.145 | -0.4595 | Yes |
| 35 | ZNF423 | NA | 47960 | -0.146 | -0.4424 | Yes |
| 36 | CCDC66 | NA | 48985 | -0.155 | -0.4420 | Yes |
| 37 | TCTN1 | NA | 49004 | -0.155 | -0.4232 | Yes |
| 38 | ZMYND10 | NA | 49557 | -0.160 | -0.4135 | Yes |
| 39 | BBS4 | NA | 49701 | -0.162 | -0.3961 | Yes |
| 40 | GGA1 | NA | 49782 | -0.163 | -0.3774 | Yes |
| 41 | TCTN2 | NA | 50286 | -0.168 | -0.3658 | Yes |
| 42 | ARL13A | NA | 50354 | -0.169 | -0.3461 | Yes |
| 43 | IFT122 | NA | 50648 | -0.173 | -0.3302 | Yes |
| 44 | BBS1 | NA | 50700 | -0.173 | -0.3097 | Yes |
| 45 | IFT140 | NA | 50735 | -0.174 | -0.2888 | Yes |
| 46 | BBS9 | NA | 50881 | -0.175 | -0.2698 | Yes |
| 47 | FAM149B1 | NA | 51027 | -0.177 | -0.2505 | Yes |
| 48 | TBC1D32 | NA | 51810 | -0.188 | -0.2415 | Yes |
| 49 | IFT80 | NA | 51971 | -0.191 | -0.2208 | Yes |
| 50 | CROCC | NA | 52070 | -0.193 | -0.1988 | Yes |
| 51 | EFCAB7 | NA | 52103 | -0.193 | -0.1755 | Yes |
| 52 | TTC21B | NA | 52143 | -0.194 | -0.1522 | Yes |
| 53 | CCDC39 | NA | 52472 | -0.200 | -0.1335 | Yes |
| 54 | CCDC88A | NA | 52582 | -0.202 | -0.1105 | Yes |
| 55 | FSIP2 | NA | 53418 | -0.221 | -0.0984 | Yes |
| 56 | SPATA7 | NA | 53486 | -0.224 | -0.0720 | Yes |
| 57 | BBIP1 | NA | 53682 | -0.231 | -0.0470 | Yes |
| 58 | TTC21A | NA | 53929 | -0.240 | -0.0219 | Yes |
| 59 | TMEM107 | NA | 54408 | -0.275 | 0.0034 | Yes |