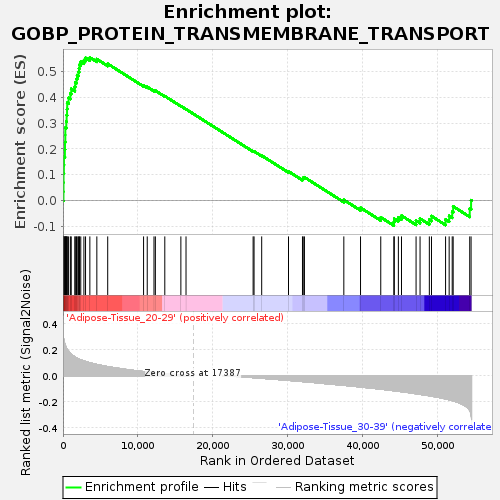
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_20-29 |
| GeneSet | GOBP_PROTEIN_TRANSMEMBRANE_TRANSPORT |
| Enrichment Score (ES) | 0.5542038 |
| Normalized Enrichment Score (NES) | 1.452808 |
| Nominal p-value | 0.09780439 |
| FDR q-value | 0.55821484 |
| FWER p-Value | 0.972 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | TIMM50 | NA | 62 | 0.294 | 0.0344 | Yes |
| 2 | TIMM23 | NA | 67 | 0.292 | 0.0696 | Yes |
| 3 | GRPEL1 | NA | 73 | 0.290 | 0.1045 | Yes |
| 4 | SEC61B | NA | 115 | 0.275 | 0.1370 | Yes |
| 5 | TOMM40 | NA | 128 | 0.271 | 0.1695 | Yes |
| 6 | SEC61G | NA | 253 | 0.244 | 0.1968 | Yes |
| 7 | AIFM1 | NA | 270 | 0.242 | 0.2257 | Yes |
| 8 | TOMM22 | NA | 308 | 0.235 | 0.2535 | Yes |
| 9 | HSPA8 | NA | 310 | 0.235 | 0.2819 | Yes |
| 10 | TIMM44 | NA | 439 | 0.216 | 0.3056 | Yes |
| 11 | SEC61A1 | NA | 486 | 0.211 | 0.3302 | Yes |
| 12 | HSPD1 | NA | 541 | 0.205 | 0.3540 | Yes |
| 13 | TIMM21 | NA | 562 | 0.203 | 0.3782 | Yes |
| 14 | TIMM17A | NA | 744 | 0.189 | 0.3977 | Yes |
| 15 | ABCA1 | NA | 986 | 0.173 | 0.4142 | Yes |
| 16 | AZGP1 | NA | 1098 | 0.165 | 0.4322 | Yes |
| 17 | PAM16 | NA | 1584 | 0.145 | 0.4408 | Yes |
| 18 | TOMM40L | NA | 1639 | 0.143 | 0.4571 | Yes |
| 19 | HSPA5 | NA | 1807 | 0.137 | 0.4706 | Yes |
| 20 | PEX5 | NA | 1891 | 0.134 | 0.4853 | Yes |
| 21 | PEX14 | NA | 2059 | 0.129 | 0.4978 | Yes |
| 22 | CHCHD4 | NA | 2145 | 0.127 | 0.5116 | Yes |
| 23 | AKT2 | NA | 2199 | 0.125 | 0.5258 | Yes |
| 24 | TIMM23B | NA | 2374 | 0.122 | 0.5373 | Yes |
| 25 | TOMM70 | NA | 2793 | 0.114 | 0.5434 | Yes |
| 26 | ROMO1 | NA | 3012 | 0.110 | 0.5527 | Yes |
| 27 | PEX10 | NA | 3588 | 0.100 | 0.5542 | Yes |
| 28 | TIMM22 | NA | 4521 | 0.087 | 0.5476 | No |
| 29 | SRP54 | NA | 5973 | 0.070 | 0.5295 | No |
| 30 | LONP2 | NA | 10765 | 0.032 | 0.4454 | No |
| 31 | TIMM17B | NA | 11255 | 0.029 | 0.4399 | No |
| 32 | TOMM20 | NA | 12157 | 0.023 | 0.4262 | No |
| 33 | DNAJC15 | NA | 12342 | 0.022 | 0.4255 | No |
| 34 | PEX16 | NA | 13601 | 0.016 | 0.4044 | No |
| 35 | DNLZ | NA | 15749 | 0.006 | 0.3658 | No |
| 36 | PEX26 | NA | 16437 | 0.004 | 0.3536 | No |
| 37 | MCL1 | NA | 25402 | -0.014 | 0.1910 | No |
| 38 | SEC63 | NA | 25527 | -0.015 | 0.1905 | No |
| 39 | PEX5L | NA | 26548 | -0.019 | 0.1741 | No |
| 40 | PEX13 | NA | 30130 | -0.035 | 0.1127 | No |
| 41 | TMED10 | NA | 31996 | -0.045 | 0.0839 | No |
| 42 | TRAM1 | NA | 32093 | -0.045 | 0.0876 | No |
| 43 | PEX7 | NA | 32259 | -0.046 | 0.0901 | No |
| 44 | GRPEL2 | NA | 37522 | -0.074 | 0.0025 | No |
| 45 | TOMM7 | NA | 39754 | -0.087 | -0.0279 | No |
| 46 | TRAM2 | NA | 42456 | -0.103 | -0.0650 | No |
| 47 | PEX2 | NA | 44205 | -0.115 | -0.0831 | No |
| 48 | SEC62 | NA | 44266 | -0.116 | -0.0702 | No |
| 49 | DNAJC19 | NA | 44810 | -0.120 | -0.0657 | No |
| 50 | TOMM20L | NA | 45227 | -0.123 | -0.0585 | No |
| 51 | BLOC1S3 | NA | 47175 | -0.138 | -0.0775 | No |
| 52 | CLU | NA | 47708 | -0.143 | -0.0699 | No |
| 53 | PEX12 | NA | 48948 | -0.154 | -0.0740 | No |
| 54 | SEC61A2 | NA | 49235 | -0.157 | -0.0603 | No |
| 55 | ZFAND2B | NA | 51117 | -0.179 | -0.0732 | No |
| 56 | PEX6 | NA | 51596 | -0.185 | -0.0596 | No |
| 57 | TRAM1L1 | NA | 52003 | -0.191 | -0.0439 | No |
| 58 | PEX1 | NA | 52132 | -0.194 | -0.0228 | No |
| 59 | RTN2 | NA | 54344 | -0.266 | -0.0311 | No |
| 60 | C2CD5 | NA | 54525 | -0.295 | 0.0012 | No |