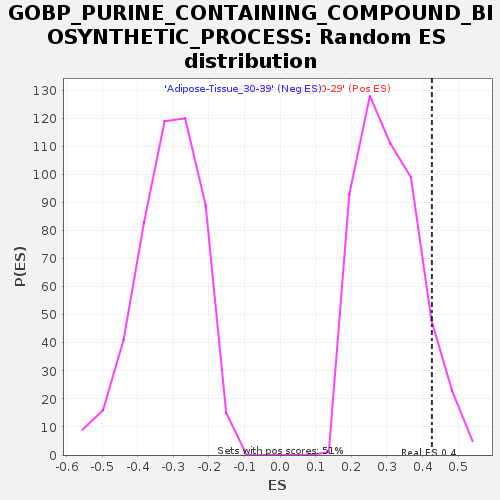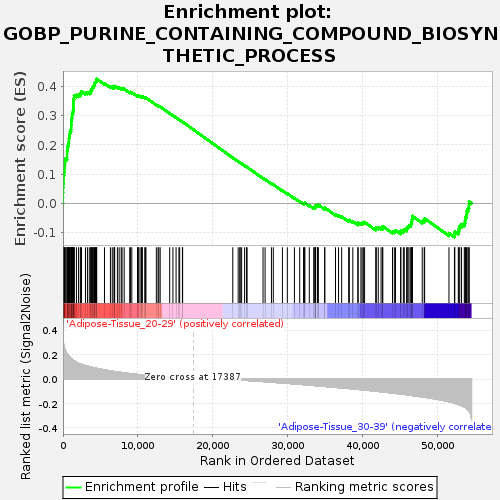
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_20-29 |
| GeneSet | GOBP_PURINE_CONTAINING_COMPOUND_BIOSYNTHETIC_PROCESS |
| Enrichment Score (ES) | 0.42614612 |
| Normalized Enrichment Score (NES) | 1.3982376 |
| Nominal p-value | 0.08858268 |
| FDR q-value | 0.6622758 |
| FWER p-Value | 0.986 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | AK2 | NA | 6 | 0.378 | 0.0187 | Yes |
| 2 | AK4 | NA | 9 | 0.365 | 0.0368 | Yes |
| 3 | GART | NA | 15 | 0.345 | 0.0538 | Yes |
| 4 | PDHA1 | NA | 43 | 0.301 | 0.0683 | Yes |
| 5 | SHMT1 | NA | 92 | 0.282 | 0.0814 | Yes |
| 6 | DLAT | NA | 94 | 0.281 | 0.0954 | Yes |
| 7 | PAICS | NA | 157 | 0.261 | 0.1072 | Yes |
| 8 | ENO1 | NA | 206 | 0.253 | 0.1188 | Yes |
| 9 | STOML2 | NA | 234 | 0.247 | 0.1306 | Yes |
| 10 | ACSL1 | NA | 252 | 0.244 | 0.1425 | Yes |
| 11 | NME1 | NA | 297 | 0.237 | 0.1534 | Yes |
| 12 | APRT | NA | 547 | 0.205 | 0.1590 | Yes |
| 13 | FASN | NA | 549 | 0.204 | 0.1692 | Yes |
| 14 | PDHX | NA | 552 | 0.204 | 0.1793 | Yes |
| 15 | ADCY3 | NA | 610 | 0.199 | 0.1881 | Yes |
| 16 | DLD | NA | 645 | 0.196 | 0.1972 | Yes |
| 17 | PFAS | NA | 735 | 0.190 | 0.2050 | Yes |
| 18 | ATIC | NA | 782 | 0.186 | 0.2134 | Yes |
| 19 | PDHB | NA | 787 | 0.186 | 0.2226 | Yes |
| 20 | ACSL4 | NA | 841 | 0.182 | 0.2306 | Yes |
| 21 | SHMT2 | NA | 897 | 0.179 | 0.2385 | Yes |
| 22 | SLC25A1 | NA | 944 | 0.176 | 0.2464 | Yes |
| 23 | MPC2 | NA | 1063 | 0.168 | 0.2526 | Yes |
| 24 | VCP | NA | 1092 | 0.166 | 0.2603 | Yes |
| 25 | DNAJC30 | NA | 1094 | 0.166 | 0.2685 | Yes |
| 26 | MTHFD1 | NA | 1119 | 0.164 | 0.2762 | Yes |
| 27 | TECR | NA | 1122 | 0.164 | 0.2843 | Yes |
| 28 | GCDH | NA | 1176 | 0.162 | 0.2914 | Yes |
| 29 | PRPS1 | NA | 1207 | 0.161 | 0.2988 | Yes |
| 30 | ACACB | NA | 1216 | 0.160 | 0.3066 | Yes |
| 31 | ALDOA | NA | 1318 | 0.156 | 0.3125 | Yes |
| 32 | HSD17B12 | NA | 1351 | 0.154 | 0.3196 | Yes |
| 33 | ADSL | NA | 1375 | 0.153 | 0.3268 | Yes |
| 34 | ACSL5 | NA | 1384 | 0.153 | 0.3342 | Yes |
| 35 | PPARA | NA | 1387 | 0.153 | 0.3418 | Yes |
| 36 | COASY | NA | 1396 | 0.152 | 0.3492 | Yes |
| 37 | PAPSS1 | NA | 1405 | 0.152 | 0.3566 | Yes |
| 38 | ACLY | NA | 1487 | 0.149 | 0.3625 | Yes |
| 39 | AMD1 | NA | 1492 | 0.149 | 0.3698 | Yes |
| 40 | PDP2 | NA | 1795 | 0.137 | 0.3711 | Yes |
| 41 | ACACA | NA | 2079 | 0.129 | 0.3722 | Yes |
| 42 | PPAT | NA | 2313 | 0.123 | 0.3741 | Yes |
| 43 | PNP | NA | 2395 | 0.121 | 0.3786 | Yes |
| 44 | IMPDH1 | NA | 2474 | 0.120 | 0.3831 | Yes |
| 45 | HACD2 | NA | 3000 | 0.110 | 0.3789 | Yes |
| 46 | ADCY8 | NA | 3265 | 0.105 | 0.3793 | Yes |
| 47 | HPRT1 | NA | 3508 | 0.101 | 0.3799 | Yes |
| 48 | ELOVL6 | NA | 3714 | 0.098 | 0.3810 | Yes |
| 49 | NPR1 | NA | 3716 | 0.098 | 0.3859 | Yes |
| 50 | ACOT7 | NA | 3823 | 0.097 | 0.3887 | Yes |
| 51 | PDK2 | NA | 3874 | 0.096 | 0.3926 | Yes |
| 52 | ELOVL1 | NA | 3911 | 0.095 | 0.3966 | Yes |
| 53 | ADCY5 | NA | 4009 | 0.094 | 0.3995 | Yes |
| 54 | PARP1 | NA | 4110 | 0.092 | 0.4023 | Yes |
| 55 | ACSS2 | NA | 4177 | 0.091 | 0.4056 | Yes |
| 56 | ELOVL5 | NA | 4225 | 0.091 | 0.4092 | Yes |
| 57 | NPPC | NA | 4234 | 0.091 | 0.4136 | Yes |
| 58 | UQCC3 | NA | 4344 | 0.089 | 0.4160 | Yes |
| 59 | ATP6V0C | NA | 4427 | 0.088 | 0.4189 | Yes |
| 60 | ELOVL3 | NA | 4468 | 0.088 | 0.4225 | Yes |
| 61 | SCD | NA | 4505 | 0.087 | 0.4261 | Yes |
| 62 | NME6 | NA | 5541 | 0.075 | 0.4108 | No |
| 63 | ACSL3 | NA | 6336 | 0.066 | 0.3995 | No |
| 64 | MLYCD | NA | 6580 | 0.064 | 0.3983 | No |
| 65 | PDK1 | NA | 6820 | 0.062 | 0.3969 | No |
| 66 | NME2 | NA | 6847 | 0.062 | 0.3995 | No |
| 67 | NOS3 | NA | 6905 | 0.061 | 0.4015 | No |
| 68 | GMPS | NA | 7273 | 0.058 | 0.3977 | No |
| 69 | PKM | NA | 7503 | 0.056 | 0.3962 | No |
| 70 | VPS9D1 | NA | 7775 | 0.054 | 0.3939 | No |
| 71 | PDHA2 | NA | 7891 | 0.053 | 0.3944 | No |
| 72 | HACD1 | NA | 8176 | 0.050 | 0.3917 | No |
| 73 | HPCA | NA | 8918 | 0.045 | 0.3803 | No |
| 74 | GUCA1A | NA | 9008 | 0.044 | 0.3809 | No |
| 75 | MTHFD2L | NA | 9194 | 0.043 | 0.3796 | No |
| 76 | MT-ATP6 | NA | 9953 | 0.037 | 0.3675 | No |
| 77 | PANK3 | NA | 9963 | 0.037 | 0.3692 | No |
| 78 | SLC35B2 | NA | 10105 | 0.036 | 0.3684 | No |
| 79 | H19 | NA | 10296 | 0.035 | 0.3667 | No |
| 80 | NPPA | NA | 10510 | 0.033 | 0.3644 | No |
| 81 | GSTZ1 | NA | 10527 | 0.033 | 0.3658 | No |
| 82 | ACSBG2 | NA | 10599 | 0.033 | 0.3661 | No |
| 83 | IMPDH2 | NA | 10924 | 0.031 | 0.3617 | No |
| 84 | NPPB | NA | 11039 | 0.030 | 0.3611 | No |
| 85 | MT-CO2 | NA | 11073 | 0.030 | 0.3620 | No |
| 86 | MTCO2P12 | NA | 12469 | 0.022 | 0.3374 | No |
| 87 | DGUOK | NA | 12697 | 0.020 | 0.3342 | No |
| 88 | ADK | NA | 12807 | 0.020 | 0.3332 | No |
| 89 | ELOVL2 | NA | 12984 | 0.019 | 0.3309 | No |
| 90 | AMPD2 | NA | 14258 | 0.013 | 0.3081 | No |
| 91 | SLC26A1 | NA | 14687 | 0.011 | 0.3008 | No |
| 92 | GUCA2B | NA | 15109 | 0.009 | 0.2935 | No |
| 93 | ACSF3 | NA | 15490 | 0.007 | 0.2869 | No |
| 94 | ACAT1 | NA | 15591 | 0.007 | 0.2854 | No |
| 95 | PRPS2 | NA | 15955 | 0.006 | 0.2790 | No |
| 96 | LDHC | NA | 15985 | 0.005 | 0.2787 | No |
| 97 | MT-ATP8 | NA | 22684 | -0.004 | 0.1559 | No |
| 98 | PANK4 | NA | 23404 | -0.007 | 0.1430 | No |
| 99 | GUCA1C | NA | 23621 | -0.008 | 0.1394 | No |
| 100 | SCD5 | NA | 23675 | -0.008 | 0.1388 | No |
| 101 | RD3 | NA | 23868 | -0.008 | 0.1357 | No |
| 102 | NME7 | NA | 24260 | -0.010 | 0.1290 | No |
| 103 | ADCY10 | NA | 24513 | -0.011 | 0.1249 | No |
| 104 | PRPS1L1 | NA | 24596 | -0.011 | 0.1240 | No |
| 105 | SLC26A2 | NA | 26726 | -0.020 | 0.0858 | No |
| 106 | NME2P1 | NA | 27002 | -0.021 | 0.0818 | No |
| 107 | NUDT2 | NA | 27850 | -0.025 | 0.0675 | No |
| 108 | NME9 | NA | 28099 | -0.026 | 0.0642 | No |
| 109 | SPHK2 | NA | 29317 | -0.032 | 0.0434 | No |
| 110 | AK1 | NA | 29973 | -0.035 | 0.0331 | No |
| 111 | PINK1 | NA | 30921 | -0.039 | 0.0177 | No |
| 112 | NME4 | NA | 31627 | -0.043 | 0.0068 | No |
| 113 | PDK3 | NA | 32138 | -0.045 | -0.0003 | No |
| 114 | IL4 | NA | 32256 | -0.046 | -0.0002 | No |
| 115 | ADCY6 | NA | 32317 | -0.046 | 0.0010 | No |
| 116 | PAPSS2 | NA | 32908 | -0.049 | -0.0074 | No |
| 117 | GUCA2A | NA | 33499 | -0.052 | -0.0156 | No |
| 118 | PDK4 | NA | 33662 | -0.053 | -0.0160 | No |
| 119 | NME5 | NA | 33680 | -0.053 | -0.0137 | No |
| 120 | ADCY2 | NA | 33694 | -0.053 | -0.0113 | No |
| 121 | PRKAG2 | NA | 33741 | -0.053 | -0.0095 | No |
| 122 | NOS1 | NA | 33748 | -0.053 | -0.0069 | No |
| 123 | AK5 | NA | 33993 | -0.055 | -0.0087 | No |
| 124 | RCVRN | NA | 34091 | -0.055 | -0.0077 | No |
| 125 | GUK1 | NA | 34094 | -0.055 | -0.0050 | No |
| 126 | PDZD3 | NA | 34971 | -0.060 | -0.0181 | No |
| 127 | PANK2 | NA | 34973 | -0.060 | -0.0152 | No |
| 128 | ADAL | NA | 36410 | -0.068 | -0.0382 | No |
| 129 | AK3 | NA | 36815 | -0.070 | -0.0422 | No |
| 130 | SLC25A13 | NA | 37233 | -0.072 | -0.0462 | No |
| 131 | FLCN | NA | 38174 | -0.077 | -0.0597 | No |
| 132 | PPT2 | NA | 38256 | -0.078 | -0.0573 | No |
| 133 | SLC35B3 | NA | 38723 | -0.080 | -0.0619 | No |
| 134 | MTAP | NA | 39394 | -0.084 | -0.0700 | No |
| 135 | ACSBG1 | NA | 39439 | -0.085 | -0.0666 | No |
| 136 | GMPR | NA | 39807 | -0.087 | -0.0690 | No |
| 137 | PPCDC | NA | 39968 | -0.088 | -0.0676 | No |
| 138 | ACSL6 | NA | 40132 | -0.089 | -0.0662 | No |
| 139 | PRPSAP1 | NA | 40296 | -0.090 | -0.0647 | No |
| 140 | ELOVL4 | NA | 41786 | -0.099 | -0.0872 | No |
| 141 | NT5E | NA | 41860 | -0.100 | -0.0836 | No |
| 142 | NOS2 | NA | 42122 | -0.101 | -0.0833 | No |
| 143 | PRPSAP2 | NA | 42500 | -0.103 | -0.0851 | No |
| 144 | AMPD1 | NA | 42706 | -0.105 | -0.0837 | No |
| 145 | PKLR | NA | 42718 | -0.105 | -0.0787 | No |
| 146 | TMSB4X | NA | 44027 | -0.114 | -0.0971 | No |
| 147 | GUCY1A2 | NA | 44269 | -0.116 | -0.0957 | No |
| 148 | HSD17B8 | NA | 44431 | -0.117 | -0.0929 | No |
| 149 | AMPD3 | NA | 45105 | -0.122 | -0.0992 | No |
| 150 | PPARGC1A | NA | 45150 | -0.122 | -0.0940 | No |
| 151 | PID1 | NA | 45478 | -0.125 | -0.0938 | No |
| 152 | GUCA1B | NA | 45595 | -0.126 | -0.0897 | No |
| 153 | ATP6V1A | NA | 45921 | -0.128 | -0.0893 | No |
| 154 | PPT1 | NA | 45967 | -0.128 | -0.0837 | No |
| 155 | PRTFDC1 | NA | 46086 | -0.129 | -0.0794 | No |
| 156 | GUCY2F | NA | 46235 | -0.131 | -0.0757 | No |
| 157 | ADCY7 | NA | 46472 | -0.132 | -0.0734 | No |
| 158 | TREM2 | NA | 46519 | -0.133 | -0.0677 | No |
| 159 | PPCS | NA | 46611 | -0.134 | -0.0627 | No |
| 160 | ADA | NA | 46625 | -0.134 | -0.0563 | No |
| 161 | GUCY2C | NA | 46659 | -0.134 | -0.0502 | No |
| 162 | PANK1 | NA | 46670 | -0.134 | -0.0438 | No |
| 163 | TGFB1 | NA | 48001 | -0.146 | -0.0609 | No |
| 164 | DCAKD | NA | 48265 | -0.148 | -0.0584 | No |
| 165 | CBR4 | NA | 48318 | -0.149 | -0.0520 | No |
| 166 | ADCY9 | NA | 51569 | -0.185 | -0.1025 | No |
| 167 | DIP2A | NA | 52300 | -0.196 | -0.1062 | No |
| 168 | ADCY4 | NA | 52350 | -0.197 | -0.0973 | No |
| 169 | NME3 | NA | 52818 | -0.206 | -0.0956 | No |
| 170 | SLC25A12 | NA | 52897 | -0.208 | -0.0867 | No |
| 171 | AK9 | NA | 53007 | -0.211 | -0.0782 | No |
| 172 | DCK | NA | 53249 | -0.217 | -0.0719 | No |
| 173 | NPR2 | NA | 53621 | -0.228 | -0.0674 | No |
| 174 | GMPR2 | NA | 53732 | -0.233 | -0.0578 | No |
| 175 | ELOVL7 | NA | 53770 | -0.234 | -0.0469 | No |
| 176 | PDPR | NA | 53928 | -0.240 | -0.0379 | No |
| 177 | PDP1 | NA | 53956 | -0.241 | -0.0264 | No |
| 178 | ADCY1 | NA | 54122 | -0.250 | -0.0170 | No |
| 179 | ACSS1 | NA | 54235 | -0.256 | -0.0063 | No |
| 180 | GUCY2D | NA | 54265 | -0.259 | 0.0060 | No |