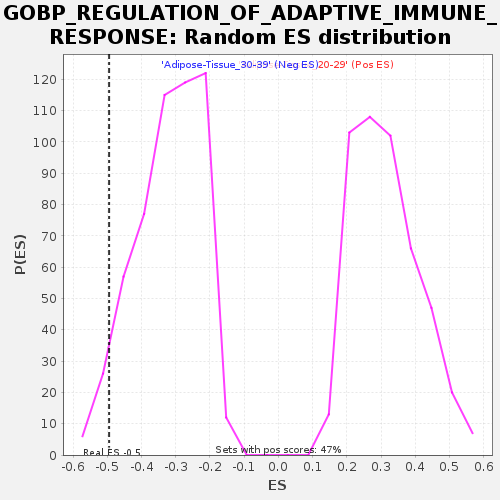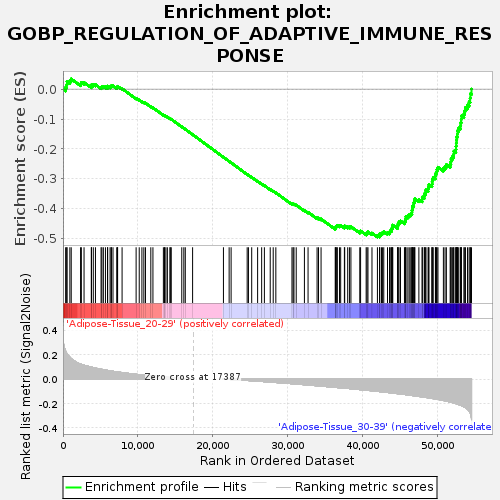
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_30-39 |
| GeneSet | GOBP_REGULATION_OF_ADAPTIVE_IMMUNE_RESPONSE |
| Enrichment Score (ES) | -0.4948498 |
| Normalized Enrichment Score (NES) | -1.5412592 |
| Nominal p-value | 0.04494382 |
| FDR q-value | 0.8567981 |
| FWER p-Value | 0.944 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | IL1RL1 | NA | 352 | 0.228 | 0.0061 | No |
| 2 | FADD | NA | 496 | 0.210 | 0.0150 | No |
| 3 | HSPD1 | NA | 541 | 0.205 | 0.0255 | No |
| 4 | IL1R1 | NA | 917 | 0.177 | 0.0283 | No |
| 5 | AZGP1 | NA | 1098 | 0.165 | 0.0341 | No |
| 6 | PVR | NA | 2347 | 0.122 | 0.0179 | No |
| 7 | CLCF1 | NA | 2453 | 0.120 | 0.0226 | No |
| 8 | PLA2G4A | NA | 2812 | 0.113 | 0.0223 | No |
| 9 | SLC22A13 | NA | 3784 | 0.097 | 0.0098 | No |
| 10 | CLC | NA | 3804 | 0.097 | 0.0147 | No |
| 11 | CD81 | NA | 4048 | 0.093 | 0.0154 | No |
| 12 | TFRC | NA | 4336 | 0.089 | 0.0150 | No |
| 13 | SMAD7 | NA | 5086 | 0.080 | 0.0057 | No |
| 14 | EXOSC3 | NA | 5196 | 0.079 | 0.0080 | No |
| 15 | ZC3H12A | NA | 5390 | 0.077 | 0.0087 | No |
| 16 | CLEC4G | NA | 5670 | 0.073 | 0.0076 | No |
| 17 | IL12A | NA | 5958 | 0.070 | 0.0062 | No |
| 18 | IL6 | NA | 5959 | 0.070 | 0.0100 | No |
| 19 | CEACAM1 | NA | 6290 | 0.067 | 0.0076 | No |
| 20 | FZD5 | NA | 6438 | 0.065 | 0.0085 | No |
| 21 | GATA3 | NA | 6465 | 0.065 | 0.0116 | No |
| 22 | ZBTB7B | NA | 6681 | 0.063 | 0.0112 | No |
| 23 | NECTIN2 | NA | 7181 | 0.059 | 0.0052 | No |
| 24 | HMCES | NA | 7278 | 0.058 | 0.0067 | No |
| 25 | MIR21 | NA | 7297 | 0.058 | 0.0095 | No |
| 26 | MAD2L2 | NA | 7894 | 0.053 | 0.0015 | No |
| 27 | ANXA1 | NA | 9769 | 0.039 | -0.0308 | No |
| 28 | NFKBIZ | NA | 10185 | 0.036 | -0.0365 | No |
| 29 | FOXJ1 | NA | 10542 | 0.033 | -0.0412 | No |
| 30 | IL23A | NA | 10803 | 0.032 | -0.0442 | No |
| 31 | C3 | NA | 11019 | 0.030 | -0.0465 | No |
| 32 | MLH1 | NA | 11733 | 0.026 | -0.0582 | No |
| 33 | ARG1 | NA | 12016 | 0.024 | -0.0621 | No |
| 34 | TRIM27 | NA | 13412 | 0.017 | -0.0868 | No |
| 35 | TNFAIP3 | NA | 13568 | 0.016 | -0.0887 | No |
| 36 | NLRP10 | NA | 13579 | 0.016 | -0.0881 | No |
| 37 | ALOX15 | NA | 13752 | 0.015 | -0.0904 | No |
| 38 | SUSD4 | NA | 13946 | 0.014 | -0.0932 | No |
| 39 | SLC11A1 | NA | 14286 | 0.013 | -0.0987 | No |
| 40 | JAK3 | NA | 14342 | 0.012 | -0.0990 | No |
| 41 | CD274 | NA | 14465 | 0.012 | -0.1006 | No |
| 42 | PAXIP1 | NA | 15869 | 0.006 | -0.1261 | No |
| 43 | CR1 | NA | 16145 | 0.005 | -0.1308 | No |
| 44 | IRF1 | NA | 16340 | 0.004 | -0.1342 | No |
| 45 | IL4R | NA | 17317 | 0.000 | -0.1521 | No |
| 46 | FUT7 | NA | 21437 | -0.000 | -0.2277 | No |
| 47 | EXOSC6 | NA | 22198 | -0.003 | -0.2415 | No |
| 48 | IL12B | NA | 22462 | -0.004 | -0.2462 | No |
| 49 | PRKCZ | NA | 24607 | -0.011 | -0.2849 | No |
| 50 | IL1B | NA | 24779 | -0.012 | -0.2874 | No |
| 51 | NFKBID | NA | 25213 | -0.013 | -0.2946 | No |
| 52 | C4BPB | NA | 26004 | -0.017 | -0.3082 | No |
| 53 | CCR2 | NA | 26543 | -0.019 | -0.3170 | No |
| 54 | IL18R1 | NA | 26908 | -0.021 | -0.3226 | No |
| 55 | NSD2 | NA | 27687 | -0.024 | -0.3356 | No |
| 56 | IL10 | NA | 28087 | -0.026 | -0.3415 | No |
| 57 | CD1D | NA | 28438 | -0.028 | -0.3464 | No |
| 58 | TRAF2 | NA | 30593 | -0.038 | -0.3839 | No |
| 59 | C4BPA | NA | 30746 | -0.038 | -0.3846 | No |
| 60 | TNFRSF1B | NA | 30859 | -0.039 | -0.3845 | No |
| 61 | IL33 | NA | 31161 | -0.040 | -0.3878 | No |
| 62 | IL4 | NA | 32256 | -0.046 | -0.4054 | No |
| 63 | TP53BP1 | NA | 32743 | -0.048 | -0.4116 | No |
| 64 | TNFSF18 | NA | 33948 | -0.054 | -0.4308 | No |
| 65 | IL23R | NA | 34139 | -0.055 | -0.4312 | No |
| 66 | SAMSN1 | NA | 34481 | -0.057 | -0.4343 | No |
| 67 | HLA-H | NA | 36372 | -0.067 | -0.4653 | No |
| 68 | NOD2 | NA | 36420 | -0.068 | -0.4625 | No |
| 69 | IFNB1 | NA | 36454 | -0.068 | -0.4593 | No |
| 70 | RIPK2 | NA | 36568 | -0.069 | -0.4576 | No |
| 71 | IL27RA | NA | 36653 | -0.069 | -0.4554 | No |
| 72 | TNF | NA | 36942 | -0.071 | -0.4568 | No |
| 73 | SLC15A4 | NA | 37101 | -0.072 | -0.4558 | No |
| 74 | CCL19 | NA | 37597 | -0.074 | -0.4608 | No |
| 75 | HLX | NA | 37639 | -0.074 | -0.4575 | No |
| 76 | FOXP3 | NA | 38039 | -0.077 | -0.4606 | No |
| 77 | EIF2AK4 | NA | 38272 | -0.078 | -0.4606 | No |
| 78 | NLRP3 | NA | 38469 | -0.079 | -0.4598 | No |
| 79 | HAVCR2 | NA | 39654 | -0.086 | -0.4768 | No |
| 80 | WAS | NA | 39762 | -0.087 | -0.4740 | No |
| 81 | DUSP10 | NA | 40505 | -0.091 | -0.4827 | No |
| 82 | IL6ST | NA | 40622 | -0.092 | -0.4798 | No |
| 83 | MSH2 | NA | 40748 | -0.092 | -0.4770 | No |
| 84 | KMT5C | NA | 41284 | -0.096 | -0.4815 | No |
| 85 | SIRT1 | NA | 42010 | -0.100 | -0.4893 | Yes |
| 86 | IL27 | NA | 42301 | -0.102 | -0.4890 | Yes |
| 87 | LTA | NA | 42318 | -0.102 | -0.4837 | Yes |
| 88 | PARP3 | NA | 42553 | -0.104 | -0.4823 | Yes |
| 89 | ZP3 | NA | 42702 | -0.105 | -0.4792 | Yes |
| 90 | CD1A | NA | 42859 | -0.106 | -0.4763 | Yes |
| 91 | SOCS5 | NA | 43359 | -0.109 | -0.4794 | Yes |
| 92 | IL20RB | NA | 43628 | -0.111 | -0.4782 | Yes |
| 93 | LILRB1 | NA | 43720 | -0.112 | -0.4738 | Yes |
| 94 | FCER2 | NA | 43885 | -0.113 | -0.4705 | Yes |
| 95 | SUPT6H | NA | 43932 | -0.113 | -0.4652 | Yes |
| 96 | PTPRC | NA | 44039 | -0.114 | -0.4608 | Yes |
| 97 | RIF1 | NA | 44053 | -0.114 | -0.4548 | Yes |
| 98 | SASH3 | NA | 44692 | -0.119 | -0.4600 | Yes |
| 99 | TNFSF4 | NA | 44738 | -0.119 | -0.4542 | Yes |
| 100 | BCL6 | NA | 44780 | -0.119 | -0.4484 | Yes |
| 101 | AGER | NA | 44895 | -0.120 | -0.4439 | Yes |
| 102 | LILRB4 | NA | 45096 | -0.122 | -0.4409 | Yes |
| 103 | MAP3K7 | NA | 45624 | -0.126 | -0.4436 | Yes |
| 104 | IRF7 | NA | 45723 | -0.127 | -0.4385 | Yes |
| 105 | CD4 | NA | 45788 | -0.127 | -0.4327 | Yes |
| 106 | CD28 | NA | 45825 | -0.127 | -0.4263 | Yes |
| 107 | IFNA2 | NA | 46081 | -0.129 | -0.4239 | Yes |
| 108 | CD48 | NA | 46261 | -0.131 | -0.4200 | Yes |
| 109 | ADCY7 | NA | 46472 | -0.132 | -0.4166 | Yes |
| 110 | IL12RB1 | NA | 46615 | -0.134 | -0.4118 | Yes |
| 111 | ADA | NA | 46625 | -0.134 | -0.4046 | Yes |
| 112 | FCGR2B | NA | 46685 | -0.134 | -0.3983 | Yes |
| 113 | MEF2C | NA | 46701 | -0.135 | -0.3912 | Yes |
| 114 | TNFSF13B | NA | 46865 | -0.136 | -0.3867 | Yes |
| 115 | CD40 | NA | 46868 | -0.136 | -0.3792 | Yes |
| 116 | IL7R | NA | 46955 | -0.137 | -0.3733 | Yes |
| 117 | IL18 | NA | 47019 | -0.137 | -0.3669 | Yes |
| 118 | PKN1 | NA | 47550 | -0.142 | -0.3689 | Yes |
| 119 | C17orf99 | NA | 47991 | -0.146 | -0.3689 | Yes |
| 120 | TGFB1 | NA | 48001 | -0.146 | -0.3610 | Yes |
| 121 | STX7 | NA | 48276 | -0.148 | -0.3579 | Yes |
| 122 | KMT5B | NA | 48286 | -0.148 | -0.3499 | Yes |
| 123 | HLA-B | NA | 48457 | -0.150 | -0.3448 | Yes |
| 124 | TNFSF13 | NA | 48487 | -0.150 | -0.3371 | Yes |
| 125 | PPP3CB | NA | 48795 | -0.153 | -0.3343 | Yes |
| 126 | HPX | NA | 48814 | -0.153 | -0.3262 | Yes |
| 127 | CD80 | NA | 48902 | -0.154 | -0.3194 | Yes |
| 128 | LOXL3 | NA | 49294 | -0.158 | -0.3179 | Yes |
| 129 | HLA-A | NA | 49322 | -0.158 | -0.3097 | Yes |
| 130 | PTPN6 | NA | 49341 | -0.158 | -0.3013 | Yes |
| 131 | KLRC1 | NA | 49468 | -0.160 | -0.2948 | Yes |
| 132 | THOC1 | NA | 49758 | -0.162 | -0.2912 | Yes |
| 133 | HMGB1 | NA | 49773 | -0.163 | -0.2825 | Yes |
| 134 | NDFIP1 | NA | 49911 | -0.164 | -0.2760 | Yes |
| 135 | HLA-G | NA | 49970 | -0.165 | -0.2680 | Yes |
| 136 | SKAP1 | NA | 50100 | -0.166 | -0.2612 | Yes |
| 137 | FBXO38 | NA | 50825 | -0.175 | -0.2649 | Yes |
| 138 | XCL1 | NA | 51042 | -0.178 | -0.2591 | Yes |
| 139 | RIPK3 | NA | 51224 | -0.180 | -0.2526 | Yes |
| 140 | RC3H2 | NA | 51730 | -0.187 | -0.2515 | Yes |
| 141 | PYCARD | NA | 51821 | -0.189 | -0.2428 | Yes |
| 142 | CD1C | NA | 51855 | -0.189 | -0.2330 | Yes |
| 143 | IL2 | NA | 52024 | -0.192 | -0.2256 | Yes |
| 144 | KLRD1 | NA | 52199 | -0.195 | -0.2180 | Yes |
| 145 | CD226 | NA | 52216 | -0.195 | -0.2076 | Yes |
| 146 | BTK | NA | 52490 | -0.200 | -0.2016 | Yes |
| 147 | RSAD2 | NA | 52512 | -0.200 | -0.1910 | Yes |
| 148 | B2M | NA | 52543 | -0.201 | -0.1805 | Yes |
| 149 | RC3H1 | NA | 52555 | -0.201 | -0.1696 | Yes |
| 150 | CD1B | NA | 52616 | -0.203 | -0.1596 | Yes |
| 151 | HLA-DRB1 | NA | 52708 | -0.204 | -0.1500 | Yes |
| 152 | HLA-F | NA | 52715 | -0.204 | -0.1389 | Yes |
| 153 | HLA-DRA | NA | 52868 | -0.208 | -0.1302 | Yes |
| 154 | TRPM4 | NA | 53090 | -0.213 | -0.1226 | Yes |
| 155 | ZBTB1 | NA | 53144 | -0.214 | -0.1118 | Yes |
| 156 | MALT1 | NA | 53209 | -0.216 | -0.1011 | Yes |
| 157 | CD160 | NA | 53232 | -0.217 | -0.0896 | Yes |
| 158 | MR1 | NA | 53563 | -0.226 | -0.0832 | Yes |
| 159 | PRKCQ | NA | 53674 | -0.231 | -0.0725 | Yes |
| 160 | HLA-E | NA | 53772 | -0.234 | -0.0614 | Yes |
| 161 | TRAF6 | NA | 54049 | -0.246 | -0.0530 | Yes |
| 162 | CD1E | NA | 54258 | -0.258 | -0.0426 | Yes |
| 163 | TBX21 | NA | 54399 | -0.273 | -0.0301 | Yes |
| 164 | HFE | NA | 54436 | -0.278 | -0.0155 | Yes |
| 165 | TNFRSF14 | NA | 54580 | -0.333 | 0.0002 | Yes |