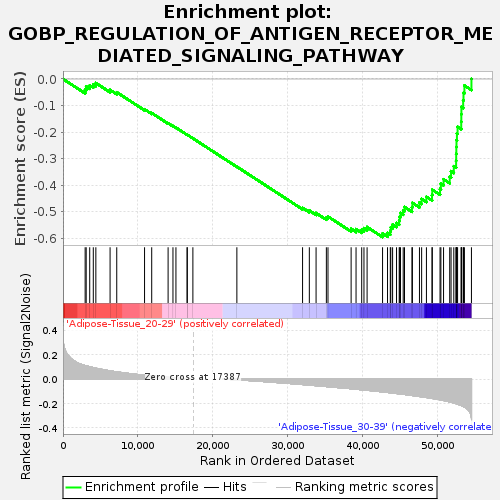
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_30-39 |
| GeneSet | GOBP_REGULATION_OF_ANTIGEN_RECEPTOR_MEDIATED_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.59633076 |
| Normalized Enrichment Score (NES) | -1.6053127 |
| Nominal p-value | 0.028571429 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.886 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | RUNX1 | NA | 2958 | 0.110 | -0.0399 | No |
| 2 | PTPN2 | NA | 3098 | 0.108 | -0.0284 | No |
| 3 | GCSAM | NA | 3570 | 0.100 | -0.0239 | No |
| 4 | CD81 | NA | 4048 | 0.093 | -0.0205 | No |
| 5 | EZR | NA | 4389 | 0.088 | -0.0153 | No |
| 6 | CEACAM1 | NA | 6290 | 0.067 | -0.0414 | No |
| 7 | NECTIN2 | NA | 7181 | 0.059 | -0.0501 | No |
| 8 | LGALS3 | NA | 10896 | 0.031 | -0.1141 | No |
| 9 | CCR7 | NA | 11853 | 0.025 | -0.1284 | No |
| 10 | RPS3 | NA | 14043 | 0.014 | -0.1667 | No |
| 11 | PAX5 | NA | 14692 | 0.011 | -0.1772 | No |
| 12 | THY1 | NA | 15090 | 0.009 | -0.1833 | No |
| 13 | PHPT1 | NA | 16581 | 0.003 | -0.2102 | No |
| 14 | PRNP | NA | 16604 | 0.003 | -0.2102 | No |
| 15 | UBASH3A | NA | 17354 | 0.000 | -0.2239 | No |
| 16 | CBFB | NA | 23231 | -0.006 | -0.3309 | No |
| 17 | NFAM1 | NA | 32009 | -0.045 | -0.4860 | No |
| 18 | RELA | NA | 32913 | -0.049 | -0.4962 | No |
| 19 | BLK | NA | 33814 | -0.054 | -0.5057 | No |
| 20 | SLC39A10 | NA | 35189 | -0.061 | -0.5229 | No |
| 21 | DUSP3 | NA | 35406 | -0.062 | -0.5188 | No |
| 22 | LYN | NA | 38497 | -0.079 | -0.5651 | No |
| 23 | DGKZ | NA | 39155 | -0.083 | -0.5664 | No |
| 24 | GPS2 | NA | 39911 | -0.088 | -0.5689 | No |
| 25 | LAPTM5 | NA | 40213 | -0.089 | -0.5628 | No |
| 26 | CD19 | NA | 40631 | -0.092 | -0.5584 | No |
| 27 | SLA2 | NA | 42698 | -0.105 | -0.5827 | Yes |
| 28 | CD300A | NA | 43361 | -0.109 | -0.5806 | Yes |
| 29 | LCK | NA | 43730 | -0.112 | -0.5728 | Yes |
| 30 | CBLB | NA | 43798 | -0.112 | -0.5594 | Yes |
| 31 | PTPRC | NA | 44039 | -0.114 | -0.5490 | Yes |
| 32 | PAWR | NA | 44565 | -0.118 | -0.5432 | Yes |
| 33 | PRKD2 | NA | 44921 | -0.121 | -0.5341 | Yes |
| 34 | GCSAML | NA | 44985 | -0.121 | -0.5195 | Yes |
| 35 | LILRB4 | NA | 45096 | -0.122 | -0.5056 | Yes |
| 36 | FCRL3 | NA | 45456 | -0.125 | -0.4960 | Yes |
| 37 | KCNN4 | NA | 45629 | -0.126 | -0.4828 | Yes |
| 38 | ADA | NA | 46625 | -0.134 | -0.4836 | Yes |
| 39 | FCGR2B | NA | 46685 | -0.134 | -0.4672 | Yes |
| 40 | RAB29 | NA | 47627 | -0.142 | -0.4659 | Yes |
| 41 | PTPRJ | NA | 47919 | -0.145 | -0.4524 | Yes |
| 42 | PTPN22 | NA | 48559 | -0.151 | -0.4445 | Yes |
| 43 | PVRIG | NA | 49306 | -0.158 | -0.4376 | Yes |
| 44 | PTPN6 | NA | 49341 | -0.158 | -0.4176 | Yes |
| 45 | GBP1 | NA | 50377 | -0.169 | -0.4146 | Yes |
| 46 | PLCL2 | NA | 50491 | -0.171 | -0.3945 | Yes |
| 47 | PRKCB | NA | 50847 | -0.175 | -0.3782 | Yes |
| 48 | CD22 | NA | 51670 | -0.186 | -0.3690 | Yes |
| 49 | CARD11 | NA | 51850 | -0.189 | -0.3477 | Yes |
| 50 | CD226 | NA | 52216 | -0.195 | -0.3290 | Yes |
| 51 | BCL10 | NA | 52525 | -0.201 | -0.3086 | Yes |
| 52 | SH2D1A | NA | 52536 | -0.201 | -0.2826 | Yes |
| 53 | RC3H1 | NA | 52555 | -0.201 | -0.2567 | Yes |
| 54 | LPXN | NA | 52575 | -0.202 | -0.2308 | Yes |
| 55 | PRKCH | NA | 52634 | -0.203 | -0.2055 | Yes |
| 56 | ELF2 | NA | 52722 | -0.204 | -0.1804 | Yes |
| 57 | MALT1 | NA | 53209 | -0.216 | -0.1613 | Yes |
| 58 | FOXP1 | NA | 53221 | -0.216 | -0.1333 | Yes |
| 59 | CD160 | NA | 53232 | -0.217 | -0.1053 | Yes |
| 60 | ELF1 | NA | 53467 | -0.223 | -0.0806 | Yes |
| 61 | TESPA1 | NA | 53512 | -0.224 | -0.0522 | Yes |
| 62 | TRAT1 | NA | 53635 | -0.229 | -0.0247 | Yes |
| 63 | STAP1 | NA | 54573 | -0.324 | 0.0003 | Yes |