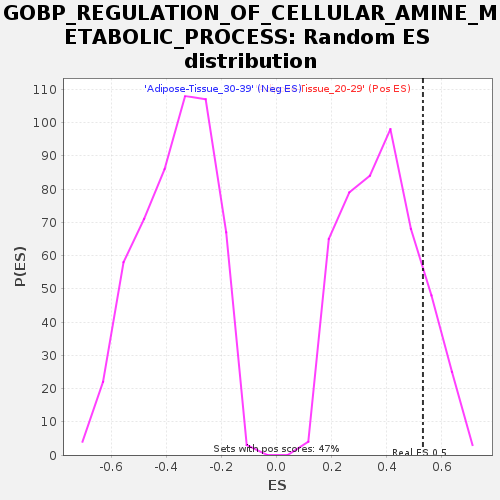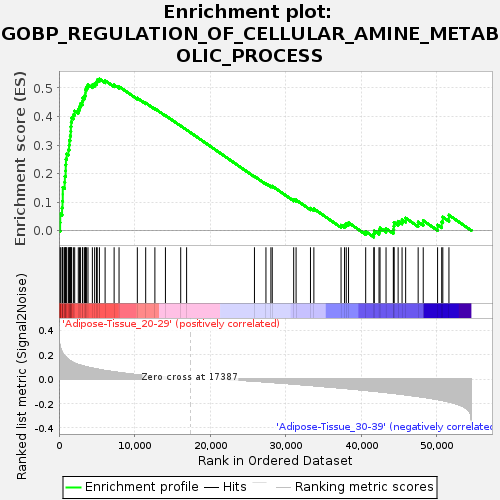
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_20-29 |
| GeneSet | GOBP_REGULATION_OF_CELLULAR_AMINE_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.531689 |
| Normalized Enrichment Score (NES) | 1.3916695 |
| Nominal p-value | 0.14767933 |
| FDR q-value | 0.67544657 |
| FWER p-Value | 0.987 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | PSME3 | NA | 147 | 0.264 | 0.0282 | Yes |
| 2 | PDE1B | NA | 162 | 0.260 | 0.0583 | Yes |
| 3 | PSMA6 | NA | 403 | 0.220 | 0.0796 | Yes |
| 4 | PSMD14 | NA | 464 | 0.213 | 0.1033 | Yes |
| 5 | PSMC4 | NA | 499 | 0.210 | 0.1272 | Yes |
| 6 | PSMA7 | NA | 500 | 0.209 | 0.1516 | Yes |
| 7 | MAOB | NA | 745 | 0.189 | 0.1693 | Yes |
| 8 | PSMB5 | NA | 777 | 0.187 | 0.1905 | Yes |
| 9 | OAZ3 | NA | 878 | 0.180 | 0.2096 | Yes |
| 10 | PSMA5 | NA | 890 | 0.179 | 0.2303 | Yes |
| 11 | PSMD8 | NA | 918 | 0.177 | 0.2506 | Yes |
| 12 | PSMB7 | NA | 1021 | 0.171 | 0.2686 | Yes |
| 13 | PSMC1 | NA | 1243 | 0.158 | 0.2831 | Yes |
| 14 | PSMD3 | NA | 1336 | 0.155 | 0.2995 | Yes |
| 15 | ODC1 | NA | 1382 | 0.153 | 0.3165 | Yes |
| 16 | PSMA2 | NA | 1480 | 0.149 | 0.3321 | Yes |
| 17 | PSMB3 | NA | 1552 | 0.147 | 0.3480 | Yes |
| 18 | PARK7 | NA | 1571 | 0.146 | 0.3646 | Yes |
| 19 | GPR37 | NA | 1614 | 0.144 | 0.3807 | Yes |
| 20 | PSMD6 | NA | 1680 | 0.141 | 0.3960 | Yes |
| 21 | SLC7A11 | NA | 1933 | 0.133 | 0.4069 | Yes |
| 22 | PSMD7 | NA | 2077 | 0.129 | 0.4193 | Yes |
| 23 | PSMB2 | NA | 2555 | 0.118 | 0.4244 | Yes |
| 24 | PSMD2 | NA | 2720 | 0.115 | 0.4348 | Yes |
| 25 | PSMB6 | NA | 2855 | 0.112 | 0.4455 | Yes |
| 26 | CHRNB2 | NA | 3116 | 0.108 | 0.4533 | Yes |
| 27 | PSMA3 | NA | 3149 | 0.107 | 0.4652 | Yes |
| 28 | PSMD9 | NA | 3385 | 0.103 | 0.4730 | Yes |
| 29 | PSMD11 | NA | 3506 | 0.101 | 0.4827 | Yes |
| 30 | HPRT1 | NA | 3508 | 0.101 | 0.4945 | Yes |
| 31 | PSMD1 | NA | 3652 | 0.099 | 0.5034 | Yes |
| 32 | PSMA4 | NA | 3838 | 0.096 | 0.5113 | Yes |
| 33 | PSMD12 | NA | 4418 | 0.088 | 0.5110 | Yes |
| 34 | PSMC2 | NA | 4717 | 0.084 | 0.5154 | Yes |
| 35 | PNKD | NA | 4959 | 0.082 | 0.5205 | Yes |
| 36 | PSMB4 | NA | 5060 | 0.080 | 0.5280 | Yes |
| 37 | PSMA1 | NA | 5351 | 0.077 | 0.5317 | Yes |
| 38 | SLC6A3 | NA | 6103 | 0.069 | 0.5259 | No |
| 39 | MIR21 | NA | 7297 | 0.058 | 0.5108 | No |
| 40 | PSMD13 | NA | 7948 | 0.052 | 0.5050 | No |
| 41 | PSMC3 | NA | 10370 | 0.034 | 0.4646 | No |
| 42 | PSMB1 | NA | 11475 | 0.027 | 0.4476 | No |
| 43 | OAZ1 | NA | 12693 | 0.020 | 0.4276 | No |
| 44 | PSMD10 | NA | 14090 | 0.014 | 0.4036 | No |
| 45 | ATCAY | NA | 16112 | 0.005 | 0.3671 | No |
| 46 | HTR1A | NA | 16892 | 0.002 | 0.3530 | No |
| 47 | ATP2B4 | NA | 25874 | -0.016 | 0.1902 | No |
| 48 | PSMF1 | NA | 27395 | -0.023 | 0.1650 | No |
| 49 | PSMC5 | NA | 28050 | -0.026 | 0.1560 | No |
| 50 | PSME4 | NA | 28230 | -0.027 | 0.1559 | No |
| 51 | NR4A2 | NA | 31067 | -0.040 | 0.1085 | No |
| 52 | PSMD4 | NA | 31385 | -0.042 | 0.1075 | No |
| 53 | PSMA8 | NA | 33290 | -0.051 | 0.0786 | No |
| 54 | PSME2 | NA | 33739 | -0.053 | 0.0766 | No |
| 55 | INS | NA | 37337 | -0.073 | 0.0191 | No |
| 56 | AZIN1 | NA | 37798 | -0.075 | 0.0194 | No |
| 57 | PSMC6 | NA | 38027 | -0.076 | 0.0242 | No |
| 58 | ACMSD | NA | 38329 | -0.078 | 0.0278 | No |
| 59 | TACR3 | NA | 40599 | -0.092 | -0.0031 | No |
| 60 | PSMB10 | NA | 41671 | -0.098 | -0.0113 | No |
| 61 | ITGAM | NA | 41735 | -0.099 | -0.0009 | No |
| 62 | PAOX | NA | 42365 | -0.103 | -0.0005 | No |
| 63 | NR1H4 | NA | 42498 | -0.103 | 0.0092 | No |
| 64 | SIRT4 | NA | 43293 | -0.109 | 0.0074 | No |
| 65 | PSMD5 | NA | 44259 | -0.116 | 0.0032 | No |
| 66 | OAZ2 | NA | 44326 | -0.116 | 0.0155 | No |
| 67 | ITGB2 | NA | 44353 | -0.116 | 0.0286 | No |
| 68 | DRD4 | NA | 44894 | -0.120 | 0.0328 | No |
| 69 | BHMT | NA | 45414 | -0.124 | 0.0378 | No |
| 70 | NQO1 | NA | 45889 | -0.128 | 0.0440 | No |
| 71 | PSMB11 | NA | 47545 | -0.142 | 0.0302 | No |
| 72 | PSME1 | NA | 48217 | -0.148 | 0.0352 | No |
| 73 | PSMB9 | NA | 50116 | -0.166 | 0.0198 | No |
| 74 | PSMB8 | NA | 50662 | -0.173 | 0.0300 | No |
| 75 | CLN3 | NA | 50809 | -0.174 | 0.0477 | No |
| 76 | SLC7A7 | NA | 51620 | -0.186 | 0.0545 | No |