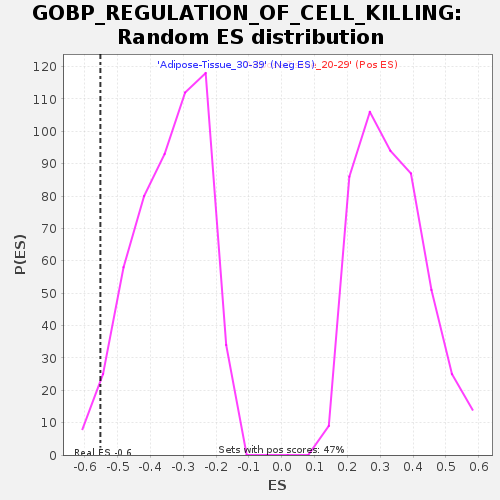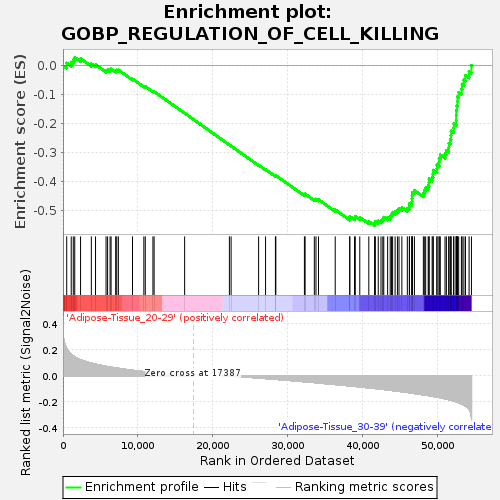
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_30-39 |
| GeneSet | GOBP_REGULATION_OF_CELL_KILLING |
| Enrichment Score (ES) | -0.5524796 |
| Normalized Enrichment Score (NES) | -1.625381 |
| Nominal p-value | 0.028409092 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.856 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | FADD | NA | 496 | 0.210 | 0.0085 | No |
| 2 | AZGP1 | NA | 1098 | 0.165 | 0.0114 | No |
| 3 | SERPINB4 | NA | 1373 | 0.153 | 0.0192 | No |
| 4 | HSP90AB1 | NA | 1567 | 0.146 | 0.0279 | No |
| 5 | PVR | NA | 2347 | 0.122 | 0.0238 | No |
| 6 | SLC22A13 | NA | 3784 | 0.097 | 0.0057 | No |
| 7 | CXCL6 | NA | 4346 | 0.089 | 0.0028 | No |
| 8 | KRT6A | NA | 5753 | 0.072 | -0.0169 | No |
| 9 | IL12A | NA | 5958 | 0.070 | -0.0148 | No |
| 10 | CEACAM1 | NA | 6290 | 0.067 | -0.0152 | No |
| 11 | CRK | NA | 6423 | 0.066 | -0.0121 | No |
| 12 | CD55 | NA | 7037 | 0.060 | -0.0183 | No |
| 13 | NECTIN2 | NA | 7181 | 0.059 | -0.0160 | No |
| 14 | IL13 | NA | 7397 | 0.057 | -0.0152 | No |
| 15 | ICAM1 | NA | 9286 | 0.042 | -0.0463 | No |
| 16 | IL23A | NA | 10803 | 0.032 | -0.0715 | No |
| 17 | LAG3 | NA | 10993 | 0.030 | -0.0724 | No |
| 18 | ARG1 | NA | 12016 | 0.024 | -0.0891 | No |
| 19 | CADM1 | NA | 12172 | 0.023 | -0.0900 | No |
| 20 | SERPINB9 | NA | 16251 | 0.004 | -0.1644 | No |
| 21 | F2RL1 | NA | 22232 | -0.003 | -0.2739 | No |
| 22 | IL12B | NA | 22462 | -0.004 | -0.2778 | No |
| 23 | BCL2L1 | NA | 26136 | -0.017 | -0.3437 | No |
| 24 | RAET1E | NA | 27064 | -0.021 | -0.3589 | No |
| 25 | IL18RAP | NA | 28382 | -0.027 | -0.3808 | No |
| 26 | CD1D | NA | 28438 | -0.028 | -0.3795 | No |
| 27 | IL4 | NA | 32256 | -0.046 | -0.4457 | No |
| 28 | CD59 | NA | 32350 | -0.046 | -0.4435 | No |
| 29 | VAV1 | NA | 33581 | -0.052 | -0.4617 | No |
| 30 | NCR1 | NA | 33801 | -0.054 | -0.4612 | No |
| 31 | IL23R | NA | 34139 | -0.055 | -0.4627 | No |
| 32 | HLA-H | NA | 36372 | -0.067 | -0.4980 | No |
| 33 | LEP | NA | 38312 | -0.078 | -0.5270 | No |
| 34 | IL21 | NA | 38317 | -0.078 | -0.5206 | No |
| 35 | CLEC12B | NA | 38954 | -0.082 | -0.5254 | No |
| 36 | CD5L | NA | 39052 | -0.082 | -0.5203 | No |
| 37 | HAVCR2 | NA | 39654 | -0.086 | -0.5241 | No |
| 38 | STAT5B | NA | 40860 | -0.093 | -0.5384 | No |
| 39 | CFH | NA | 41631 | -0.098 | -0.5443 | Yes |
| 40 | ITGAM | NA | 41735 | -0.099 | -0.5379 | Yes |
| 41 | NOS2 | NA | 42122 | -0.101 | -0.5364 | Yes |
| 42 | AP1G1 | NA | 42488 | -0.103 | -0.5345 | Yes |
| 43 | NCR3 | NA | 42731 | -0.105 | -0.5301 | Yes |
| 44 | CD1A | NA | 42859 | -0.106 | -0.5235 | Yes |
| 45 | DNASE1L3 | NA | 43377 | -0.109 | -0.5238 | Yes |
| 46 | LILRB1 | NA | 43720 | -0.112 | -0.5207 | Yes |
| 47 | FCER2 | NA | 43885 | -0.113 | -0.5142 | Yes |
| 48 | PTPRC | NA | 44039 | -0.114 | -0.5075 | Yes |
| 49 | CR1L | NA | 44373 | -0.116 | -0.5038 | Yes |
| 50 | CASP8 | NA | 44685 | -0.119 | -0.4995 | Yes |
| 51 | AGER | NA | 44895 | -0.120 | -0.4933 | Yes |
| 52 | DNASE1 | NA | 45277 | -0.123 | -0.4899 | Yes |
| 53 | ARRB2 | NA | 46006 | -0.129 | -0.4925 | Yes |
| 54 | LGALS9 | NA | 46274 | -0.131 | -0.4864 | Yes |
| 55 | TYROBP | NA | 46299 | -0.131 | -0.4758 | Yes |
| 56 | IL12RB1 | NA | 46615 | -0.134 | -0.4704 | Yes |
| 57 | RASGRP1 | NA | 46632 | -0.134 | -0.4594 | Yes |
| 58 | KLRC2 | NA | 46646 | -0.134 | -0.4484 | Yes |
| 59 | FCGR2B | NA | 46685 | -0.134 | -0.4378 | Yes |
| 60 | IL7R | NA | 46955 | -0.137 | -0.4313 | Yes |
| 61 | PIK3R6 | NA | 48148 | -0.147 | -0.4408 | Yes |
| 62 | STX7 | NA | 48276 | -0.148 | -0.4307 | Yes |
| 63 | HLA-B | NA | 48457 | -0.150 | -0.4214 | Yes |
| 64 | PPP3CB | NA | 48795 | -0.153 | -0.4148 | Yes |
| 65 | KIR2DL4 | NA | 48900 | -0.154 | -0.4038 | Yes |
| 66 | SYK | NA | 48916 | -0.154 | -0.3911 | Yes |
| 67 | HLA-A | NA | 49322 | -0.158 | -0.3853 | Yes |
| 68 | BAD | NA | 49440 | -0.159 | -0.3741 | Yes |
| 69 | KLRC1 | NA | 49468 | -0.160 | -0.3611 | Yes |
| 70 | CLEC7A | NA | 49956 | -0.165 | -0.3563 | Yes |
| 71 | HLA-G | NA | 49970 | -0.165 | -0.3427 | Yes |
| 72 | CRTAM | NA | 50250 | -0.168 | -0.3337 | Yes |
| 73 | SLAMF6 | NA | 50263 | -0.168 | -0.3198 | Yes |
| 74 | LAMP1 | NA | 50411 | -0.170 | -0.3083 | Yes |
| 75 | XCL1 | NA | 51042 | -0.178 | -0.3050 | Yes |
| 76 | RIPK3 | NA | 51224 | -0.180 | -0.2932 | Yes |
| 77 | PRF1 | NA | 51544 | -0.185 | -0.2835 | Yes |
| 78 | BCL2L11 | NA | 51558 | -0.185 | -0.2683 | Yes |
| 79 | MICA | NA | 51737 | -0.187 | -0.2558 | Yes |
| 80 | P2RX7 | NA | 51834 | -0.189 | -0.2417 | Yes |
| 81 | CD1C | NA | 51855 | -0.189 | -0.2262 | Yes |
| 82 | KLRD1 | NA | 52199 | -0.195 | -0.2162 | Yes |
| 83 | CD226 | NA | 52216 | -0.195 | -0.2001 | Yes |
| 84 | SH2D1A | NA | 52536 | -0.201 | -0.1891 | Yes |
| 85 | B2M | NA | 52543 | -0.201 | -0.1723 | Yes |
| 86 | IFNG | NA | 52552 | -0.201 | -0.1556 | Yes |
| 87 | CD1B | NA | 52616 | -0.203 | -0.1397 | Yes |
| 88 | HLA-DRB1 | NA | 52708 | -0.204 | -0.1242 | Yes |
| 89 | HLA-F | NA | 52715 | -0.204 | -0.1072 | Yes |
| 90 | HLA-DRA | NA | 52868 | -0.208 | -0.0926 | Yes |
| 91 | CD160 | NA | 53232 | -0.217 | -0.0810 | Yes |
| 92 | KLRK1 | NA | 53328 | -0.219 | -0.0644 | Yes |
| 93 | MR1 | NA | 53563 | -0.226 | -0.0497 | Yes |
| 94 | HLA-E | NA | 53772 | -0.234 | -0.0339 | Yes |
| 95 | CD1E | NA | 54258 | -0.258 | -0.0211 | Yes |
| 96 | STAP1 | NA | 54573 | -0.324 | 0.0003 | Yes |