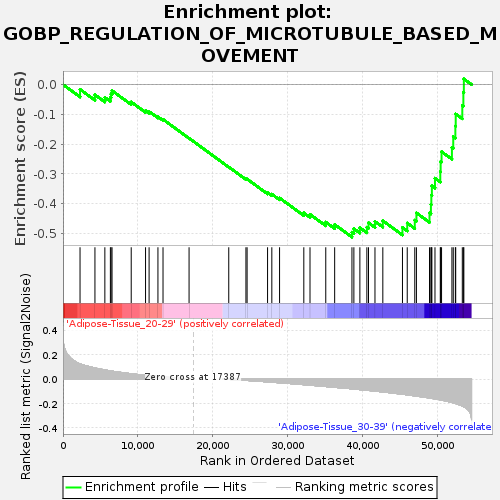
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_30-39 |
| GeneSet | GOBP_REGULATION_OF_MICROTUBULE_BASED_MOVEMENT |
| Enrichment Score (ES) | -0.51212704 |
| Normalized Enrichment Score (NES) | -1.550656 |
| Nominal p-value | 0.011904762 |
| FDR q-value | 0.86071205 |
| FWER p-Value | 0.936 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | RNASE10 | NA | 2276 | 0.124 | -0.0162 | No |
| 2 | EPPIN | NA | 4267 | 0.090 | -0.0342 | No |
| 3 | DRC1 | NA | 5592 | 0.074 | -0.0433 | No |
| 4 | GAS2L2 | NA | 6330 | 0.066 | -0.0431 | No |
| 5 | CNIH2 | NA | 6419 | 0.066 | -0.0313 | No |
| 6 | PGAM4 | NA | 6551 | 0.064 | -0.0204 | No |
| 7 | NEFH | NA | 9114 | 0.043 | -0.0585 | No |
| 8 | CFAP69 | NA | 11029 | 0.030 | -0.0874 | No |
| 9 | PRDM14 | NA | 11503 | 0.027 | -0.0905 | No |
| 10 | CFAP20 | NA | 12685 | 0.020 | -0.1080 | No |
| 11 | DNAH11 | NA | 13364 | 0.017 | -0.1169 | No |
| 12 | STK11 | NA | 16844 | 0.002 | -0.1802 | No |
| 13 | DEFB1 | NA | 22147 | -0.003 | -0.2768 | No |
| 14 | CCDC65 | NA | 24442 | -0.011 | -0.3167 | No |
| 15 | ARMC4 | NA | 24598 | -0.011 | -0.3172 | No |
| 16 | TEX101 | NA | 27333 | -0.023 | -0.3627 | No |
| 17 | TPPP2 | NA | 27902 | -0.025 | -0.3679 | No |
| 18 | IQCF1 | NA | 28936 | -0.030 | -0.3807 | No |
| 19 | MKKS | NA | 32176 | -0.045 | -0.4308 | No |
| 20 | CALY | NA | 33003 | -0.050 | -0.4357 | No |
| 21 | TRIM46 | NA | 35109 | -0.061 | -0.4618 | No |
| 22 | CFAP43 | NA | 36298 | -0.067 | -0.4698 | No |
| 23 | HAP1 | NA | 38607 | -0.080 | -0.4957 | Yes |
| 24 | DNAAF1 | NA | 38865 | -0.081 | -0.4838 | Yes |
| 25 | CCDC40 | NA | 39667 | -0.086 | -0.4808 | Yes |
| 26 | TACR3 | NA | 40599 | -0.092 | -0.4790 | Yes |
| 27 | SEMG2 | NA | 40819 | -0.093 | -0.4639 | Yes |
| 28 | SEMG1 | NA | 41683 | -0.098 | -0.4595 | Yes |
| 29 | RHOT1 | NA | 42733 | -0.105 | -0.4571 | Yes |
| 30 | CCR6 | NA | 45357 | -0.124 | -0.4798 | Yes |
| 31 | RGN | NA | 45995 | -0.129 | -0.4650 | Yes |
| 32 | BORCS5 | NA | 47002 | -0.137 | -0.4553 | Yes |
| 33 | IGBP1 | NA | 47222 | -0.139 | -0.4308 | Yes |
| 34 | TAC4 | NA | 48971 | -0.155 | -0.4310 | Yes |
| 35 | TACR2 | NA | 49159 | -0.156 | -0.4023 | Yes |
| 36 | CATSPER1 | NA | 49215 | -0.157 | -0.3711 | Yes |
| 37 | TACR1 | NA | 49287 | -0.158 | -0.3400 | Yes |
| 38 | BBS4 | NA | 49701 | -0.162 | -0.3143 | Yes |
| 39 | LAMP1 | NA | 50411 | -0.170 | -0.2925 | Yes |
| 40 | TTLL6 | NA | 50459 | -0.170 | -0.2584 | Yes |
| 41 | HDAC6 | NA | 50580 | -0.172 | -0.2253 | Yes |
| 42 | FEZ1 | NA | 51965 | -0.191 | -0.2114 | Yes |
| 43 | TTC21B | NA | 52143 | -0.194 | -0.1748 | Yes |
| 44 | BBS2 | NA | 52424 | -0.199 | -0.1391 | Yes |
| 45 | CCDC39 | NA | 52472 | -0.200 | -0.0989 | Yes |
| 46 | MAP2 | NA | 53352 | -0.220 | -0.0698 | Yes |
| 47 | CFAP206 | NA | 53495 | -0.224 | -0.0264 | Yes |
| 48 | CCSAP | NA | 53559 | -0.226 | 0.0189 | Yes |