Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_20-29 |
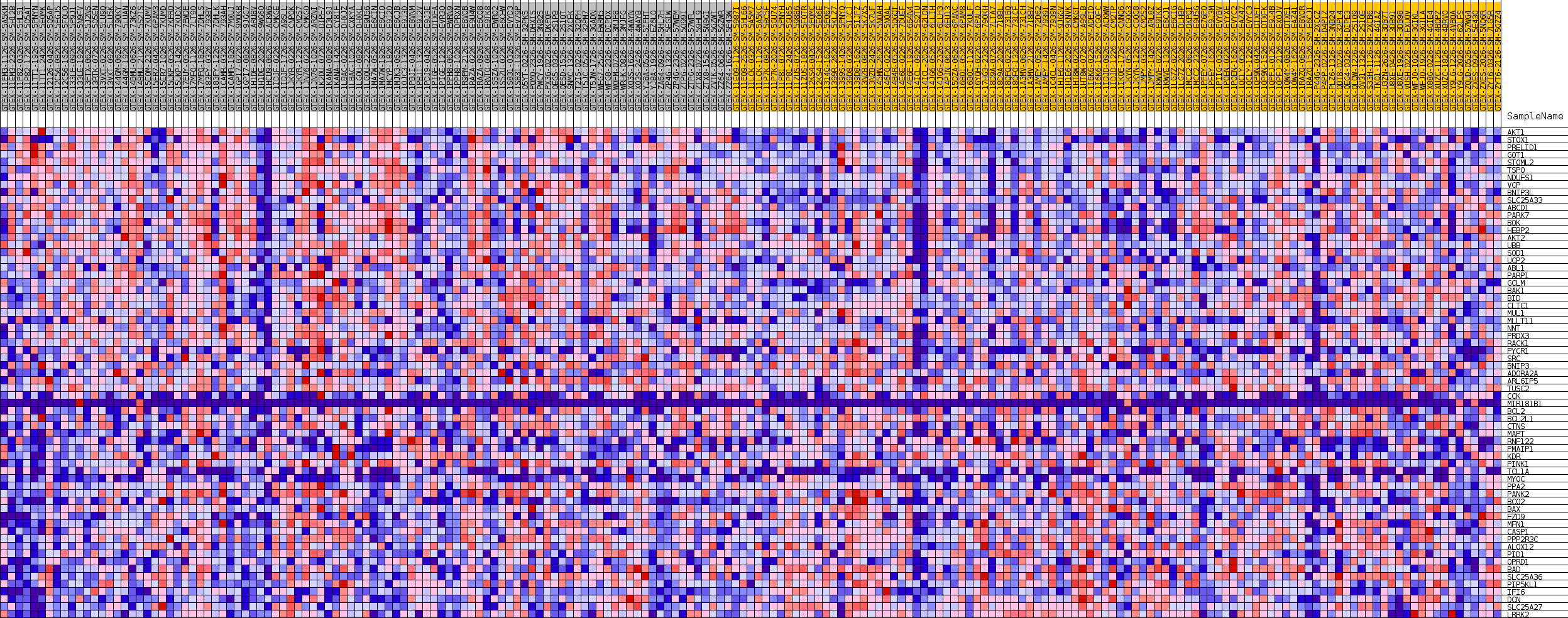
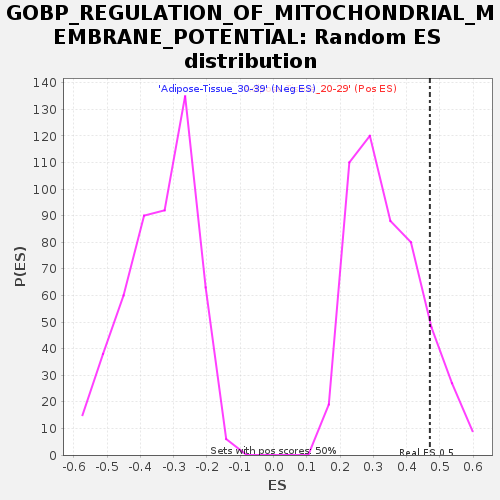
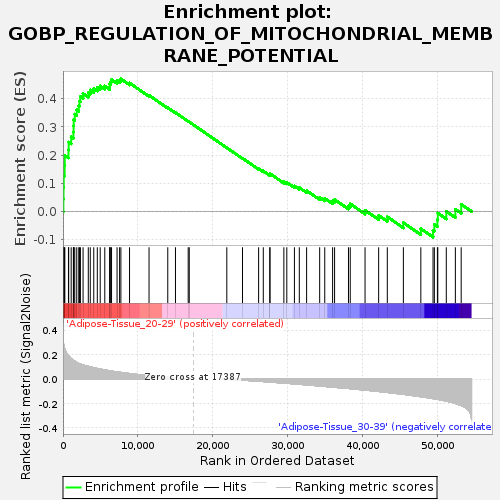
| GeneSet | GOBP_REGULATION_OF_MITOCHONDRIAL_MEMBRANE_POTENTIAL |
| Enrichment Score (ES) | 0.4699601 |
| Normalized Enrichment Score (NES) | 1.3914678 |
| Nominal p-value | 0.119760476 |
| FDR q-value | 0.6691952 |
| FWER p-Value | 0.987 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | AKT1 | NA | 49 | 0.299 | 0.0436 | Yes |
| 2 | STOX1 | NA | 70 | 0.290 | 0.0864 | Yes |
| 3 | PRELID1 | NA | 99 | 0.279 | 0.1274 | Yes |
| 4 | GOT1 | NA | 216 | 0.251 | 0.1626 | Yes |
| 5 | STOML2 | NA | 234 | 0.247 | 0.1990 | Yes |
| 6 | TSPO | NA | 729 | 0.190 | 0.2182 | Yes |
| 7 | NDUFS1 | NA | 761 | 0.188 | 0.2456 | Yes |
| 8 | VCP | NA | 1092 | 0.166 | 0.2643 | Yes |
| 9 | BNIP3L | NA | 1385 | 0.153 | 0.2816 | Yes |
| 10 | SLC25A33 | NA | 1416 | 0.152 | 0.3036 | Yes |
| 11 | ABCD1 | NA | 1427 | 0.151 | 0.3259 | Yes |
| 12 | PARK7 | NA | 1571 | 0.146 | 0.3449 | Yes |
| 13 | BOK | NA | 1838 | 0.136 | 0.3603 | Yes |
| 14 | HEBP2 | NA | 2108 | 0.128 | 0.3743 | Yes |
| 15 | AKT2 | NA | 2199 | 0.125 | 0.3913 | Yes |
| 16 | UBB | NA | 2328 | 0.123 | 0.4073 | Yes |
| 17 | SOD1 | NA | 2681 | 0.116 | 0.4180 | Yes |
| 18 | UCP2 | NA | 3379 | 0.103 | 0.4206 | Yes |
| 19 | ABL1 | NA | 3645 | 0.099 | 0.4305 | Yes |
| 20 | PARP1 | NA | 4110 | 0.092 | 0.4357 | Yes |
| 21 | GCLM | NA | 4584 | 0.086 | 0.4399 | Yes |
| 22 | BAK1 | NA | 4972 | 0.081 | 0.4449 | Yes |
| 23 | BID | NA | 5583 | 0.074 | 0.4447 | Yes |
| 24 | CLIC1 | NA | 6244 | 0.067 | 0.4426 | Yes |
| 25 | MUL1 | NA | 6247 | 0.067 | 0.4526 | Yes |
| 26 | MLLT11 | NA | 6371 | 0.066 | 0.4602 | Yes |
| 27 | NNT | NA | 6486 | 0.065 | 0.4678 | Yes |
| 28 | PRDX3 | NA | 7223 | 0.059 | 0.4630 | Yes |
| 29 | RACK1 | NA | 7561 | 0.056 | 0.4651 | Yes |
| 30 | PYCR1 | NA | 7734 | 0.054 | 0.4700 | Yes |
| 31 | SRC | NA | 8890 | 0.045 | 0.4555 | No |
| 32 | BNIP3 | NA | 11495 | 0.027 | 0.4117 | No |
| 33 | ADORA2A | NA | 13999 | 0.014 | 0.3679 | No |
| 34 | ARL6IP5 | NA | 15027 | 0.009 | 0.3505 | No |
| 35 | TUSC2 | NA | 16727 | 0.003 | 0.3197 | No |
| 36 | CCK | NA | 16869 | 0.002 | 0.3175 | No |
| 37 | MIR181B1 | NA | 21888 | -0.002 | 0.2258 | No |
| 38 | BCL2 | NA | 23983 | -0.009 | 0.1887 | No |
| 39 | BCL2L1 | NA | 26136 | -0.017 | 0.1518 | No |
| 40 | CTNS | NA | 26761 | -0.020 | 0.1434 | No |
| 41 | MAPT | NA | 27641 | -0.024 | 0.1308 | No |
| 42 | RNF122 | NA | 27648 | -0.024 | 0.1343 | No |
| 43 | PMAIP1 | NA | 29511 | -0.033 | 0.1050 | No |
| 44 | KDR | NA | 29904 | -0.035 | 0.1030 | No |
| 45 | PINK1 | NA | 30921 | -0.039 | 0.0901 | No |
| 46 | TCL1A | NA | 31574 | -0.042 | 0.0845 | No |
| 47 | MYOC | NA | 32550 | -0.047 | 0.0736 | No |
| 48 | PPA2 | NA | 34295 | -0.056 | 0.0500 | No |
| 49 | PANK2 | NA | 34973 | -0.060 | 0.0466 | No |
| 50 | BCO2 | NA | 36023 | -0.066 | 0.0371 | No |
| 51 | BAX | NA | 36287 | -0.067 | 0.0422 | No |
| 52 | FZD9 | NA | 38143 | -0.077 | 0.0197 | No |
| 53 | MFN1 | NA | 38380 | -0.078 | 0.0270 | No |
| 54 | CASP1 | NA | 40358 | -0.090 | 0.0041 | No |
| 55 | PPP2R3C | NA | 42176 | -0.102 | -0.0141 | No |
| 56 | ALOX12 | NA | 43331 | -0.109 | -0.0190 | No |
| 57 | PID1 | NA | 45478 | -0.125 | -0.0398 | No |
| 58 | OPRD1 | NA | 47811 | -0.144 | -0.0611 | No |
| 59 | BAD | NA | 49440 | -0.159 | -0.0673 | No |
| 60 | SLC25A36 | NA | 49620 | -0.161 | -0.0466 | No |
| 61 | PIP5KL1 | NA | 50024 | -0.165 | -0.0294 | No |
| 62 | IFI6 | NA | 50079 | -0.166 | -0.0057 | No |
| 63 | DCN | NA | 51213 | -0.180 | 0.0002 | No |
| 64 | SLC25A27 | NA | 52425 | -0.199 | 0.0076 | No |
| 65 | LRRK2 | NA | 53203 | -0.216 | 0.0255 | No |