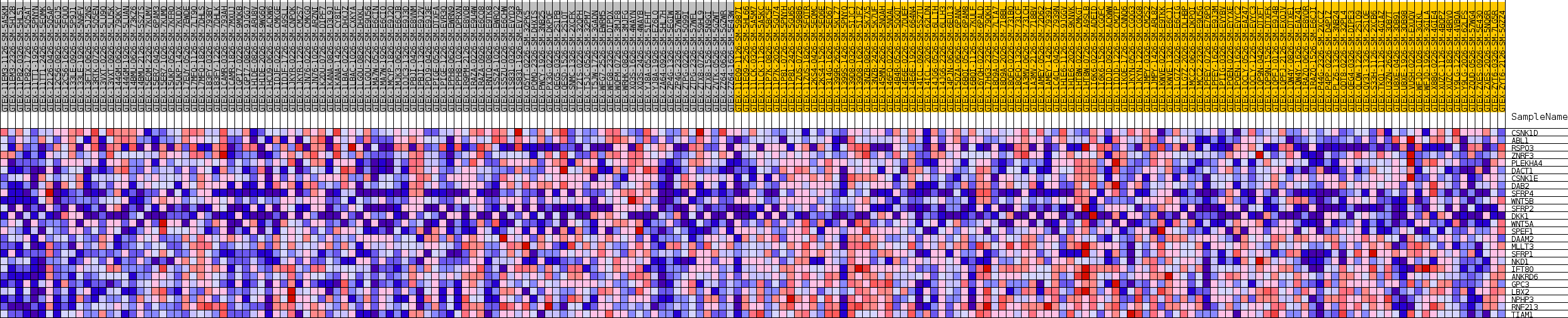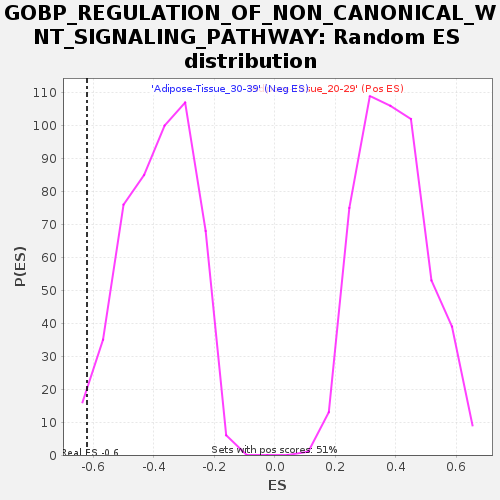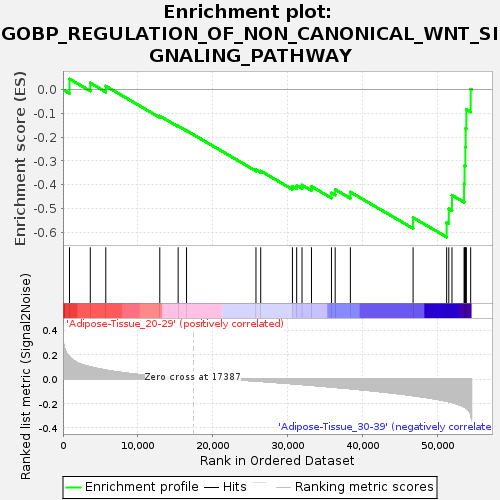
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_30-39 |
| GeneSet | GOBP_REGULATION_OF_NON_CANONICAL_WNT_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.62041867 |
| Normalized Enrichment Score (NES) | -1.6121986 |
| Nominal p-value | 0.014198783 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.876 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | CSNK1D | NA | 868 | 0.180 | 0.0454 | No |
| 2 | ABL1 | NA | 3645 | 0.099 | 0.0283 | No |
| 3 | RSPO3 | NA | 5716 | 0.073 | 0.0151 | No |
| 4 | ZNRF3 | NA | 12932 | 0.019 | -0.1106 | No |
| 5 | PLEKHA4 | NA | 15384 | 0.008 | -0.1529 | No |
| 6 | DACT1 | NA | 16507 | 0.004 | -0.1723 | No |
| 7 | CSNK1E | NA | 25782 | -0.016 | -0.3368 | No |
| 8 | DAB2 | NA | 26417 | -0.019 | -0.3421 | No |
| 9 | SFRP4 | NA | 30655 | -0.038 | -0.4068 | No |
| 10 | WNT5B | NA | 31221 | -0.041 | -0.4033 | No |
| 11 | SFRP2 | NA | 31935 | -0.044 | -0.4013 | No |
| 12 | DKK1 | NA | 33199 | -0.050 | -0.4073 | No |
| 13 | WNT5A | NA | 35870 | -0.065 | -0.4342 | No |
| 14 | SPEF1 | NA | 36366 | -0.067 | -0.4203 | No |
| 15 | DAAM2 | NA | 38398 | -0.079 | -0.4307 | No |
| 16 | MLLT3 | NA | 46777 | -0.135 | -0.5383 | No |
| 17 | SFRP1 | NA | 51261 | -0.180 | -0.5590 | Yes |
| 18 | NKD1 | NA | 51546 | -0.185 | -0.5013 | Yes |
| 19 | IFT80 | NA | 51971 | -0.191 | -0.4441 | Yes |
| 20 | ANKRD6 | NA | 53590 | -0.227 | -0.3964 | Yes |
| 21 | GPC3 | NA | 53654 | -0.230 | -0.3194 | Yes |
| 22 | LBX2 | NA | 53760 | -0.233 | -0.2418 | Yes |
| 23 | NPHP3 | NA | 53797 | -0.234 | -0.1626 | Yes |
| 24 | RNF213 | NA | 53895 | -0.238 | -0.0833 | Yes |
| 25 | TIAM1 | NA | 54471 | -0.282 | 0.0022 | Yes |