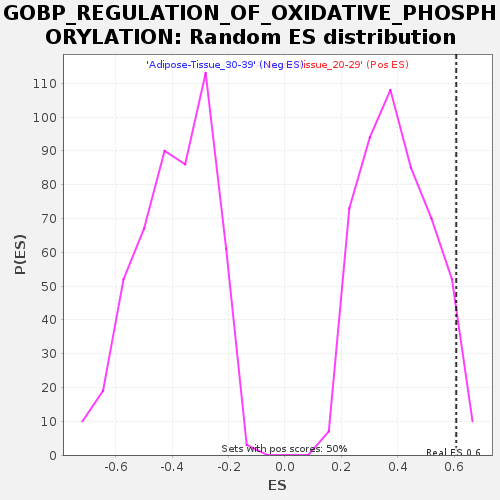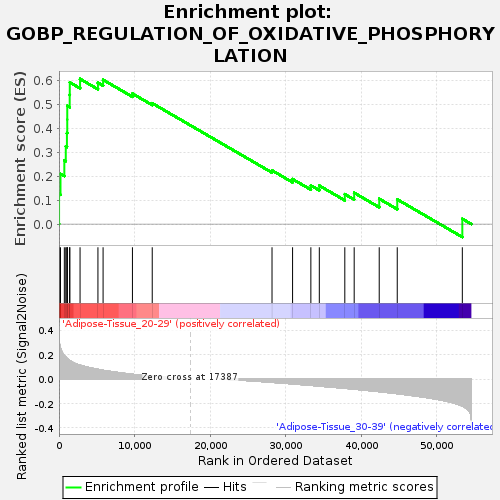
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_20-29 |
| GeneSet | GOBP_REGULATION_OF_OXIDATIVE_PHOSPHORYLATION |
| Enrichment Score (ES) | 0.60786813 |
| Normalized Enrichment Score (NES) | 1.5320317 |
| Nominal p-value | 0.044088177 |
| FDR q-value | 0.45351538 |
| FWER p-Value | 0.936 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | AK4 | NA | 9 | 0.365 | 0.1262 | Yes |
| 2 | PPIF | NA | 197 | 0.253 | 0.2103 | Yes |
| 3 | COX7A2 | NA | 706 | 0.192 | 0.2673 | Yes |
| 4 | SHMT2 | NA | 897 | 0.179 | 0.3256 | Yes |
| 5 | PDE12 | NA | 1049 | 0.169 | 0.3811 | Yes |
| 6 | VCP | NA | 1092 | 0.166 | 0.4377 | Yes |
| 7 | DNAJC30 | NA | 1094 | 0.166 | 0.4949 | Yes |
| 8 | SLC25A33 | NA | 1416 | 0.152 | 0.5414 | Yes |
| 9 | ABCD1 | NA | 1427 | 0.151 | 0.5934 | Yes |
| 10 | MLXIPL | NA | 2787 | 0.114 | 0.6079 | Yes |
| 11 | UQCC2 | NA | 5154 | 0.079 | 0.5919 | No |
| 12 | MYOG | NA | 5836 | 0.071 | 0.6041 | No |
| 13 | COX7A2P2 | NA | 9721 | 0.039 | 0.5464 | No |
| 14 | DNAJC15 | NA | 12342 | 0.022 | 0.5060 | No |
| 15 | SLC25A23 | NA | 28199 | -0.027 | 0.2247 | No |
| 16 | PINK1 | NA | 30921 | -0.039 | 0.1883 | No |
| 17 | MIR210 | NA | 33341 | -0.051 | 0.1617 | No |
| 18 | COX7A1 | NA | 34460 | -0.057 | 0.1610 | No |
| 19 | COX7A2L | NA | 37835 | -0.075 | 0.1252 | No |
| 20 | SNCA | NA | 39076 | -0.083 | 0.1310 | No |
| 21 | ACTN3 | NA | 42391 | -0.103 | 0.1058 | No |
| 22 | NUPR1 | NA | 44777 | -0.119 | 0.1034 | No |
| 23 | ATP7A | NA | 53392 | -0.221 | 0.0220 | No |