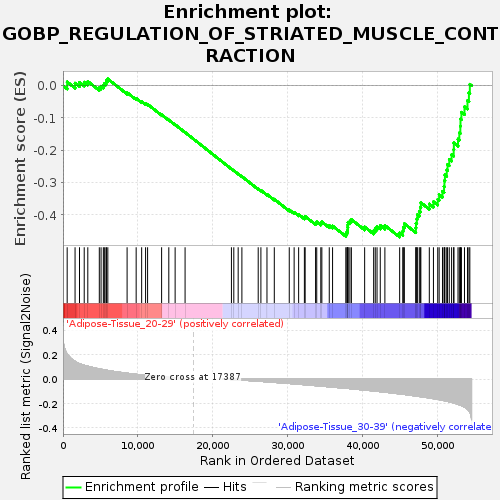
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_30-39 |
| GeneSet | GOBP_REGULATION_OF_STRIATED_MUSCLE_CONTRACTION |
| Enrichment Score (ES) | -0.468739 |
| Normalized Enrichment Score (NES) | -1.5368977 |
| Nominal p-value | 0.027613413 |
| FDR q-value | 0.8200134 |
| FWER p-Value | 0.946 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | HCN4 | NA | 557 | 0.204 | 0.0116 | No |
| 2 | PRKACA | NA | 1611 | 0.144 | 0.0077 | No |
| 3 | ADORA1 | NA | 2204 | 0.125 | 0.0102 | No |
| 4 | ATP2A2 | NA | 2835 | 0.113 | 0.0108 | No |
| 5 | BIN1 | NA | 3314 | 0.105 | 0.0132 | No |
| 6 | CHGA | NA | 4851 | 0.083 | -0.0061 | No |
| 7 | SMAD7 | NA | 5086 | 0.080 | -0.0018 | No |
| 8 | ZC3H12A | NA | 5390 | 0.077 | 0.0008 | No |
| 9 | SLC9A1 | NA | 5524 | 0.075 | 0.0064 | No |
| 10 | PKP2 | NA | 5741 | 0.072 | 0.0102 | No |
| 11 | FGF13 | NA | 5802 | 0.072 | 0.0167 | No |
| 12 | ATP1A1 | NA | 5989 | 0.070 | 0.0208 | No |
| 13 | GJA5 | NA | 8564 | 0.047 | -0.0214 | No |
| 14 | SCN4A | NA | 9775 | 0.039 | -0.0394 | No |
| 15 | NPPA | NA | 10510 | 0.033 | -0.0493 | No |
| 16 | BMP10 | NA | 11034 | 0.030 | -0.0557 | No |
| 17 | STC1 | NA | 11303 | 0.028 | -0.0576 | No |
| 18 | MYBPH | NA | 13176 | 0.018 | -0.0900 | No |
| 19 | MIR30E | NA | 14137 | 0.013 | -0.1062 | No |
| 20 | GATA4 | NA | 14982 | 0.009 | -0.1207 | No |
| 21 | TNNI3 | NA | 16311 | 0.004 | -0.1446 | No |
| 22 | ATP1A2 | NA | 22504 | -0.004 | -0.2577 | No |
| 23 | ANK2 | NA | 22821 | -0.005 | -0.2630 | No |
| 24 | ATP2A1 | NA | 23405 | -0.007 | -0.2730 | No |
| 25 | ACE2 | NA | 23902 | -0.009 | -0.2812 | No |
| 26 | MIR1-1 | NA | 26082 | -0.017 | -0.3193 | No |
| 27 | PDE4D | NA | 26442 | -0.019 | -0.3239 | No |
| 28 | RGS2 | NA | 27253 | -0.022 | -0.3364 | No |
| 29 | DSP | NA | 28232 | -0.027 | -0.3514 | No |
| 30 | MYBPC3 | NA | 30231 | -0.036 | -0.3843 | No |
| 31 | CALM1 | NA | 30883 | -0.039 | -0.3920 | No |
| 32 | NKX2-5 | NA | 31484 | -0.042 | -0.3985 | No |
| 33 | CAMK2D | NA | 32252 | -0.046 | -0.4077 | No |
| 34 | ATP1B1 | NA | 32355 | -0.046 | -0.4046 | No |
| 35 | NOS1 | NA | 33748 | -0.053 | -0.4245 | No |
| 36 | SRI | NA | 33903 | -0.054 | -0.4215 | No |
| 37 | HDAC4 | NA | 34445 | -0.057 | -0.4253 | No |
| 38 | SCN5A | NA | 34586 | -0.058 | -0.4217 | No |
| 39 | GSTO1 | NA | 35576 | -0.063 | -0.4330 | No |
| 40 | EHD3 | NA | 36018 | -0.066 | -0.4341 | No |
| 41 | SCN10A | NA | 37788 | -0.075 | -0.4585 | Yes |
| 42 | DSG2 | NA | 37924 | -0.076 | -0.4529 | Yes |
| 43 | ADRA1A | NA | 38024 | -0.076 | -0.4465 | Yes |
| 44 | DSC2 | NA | 38030 | -0.076 | -0.4384 | Yes |
| 45 | UCN | NA | 38063 | -0.077 | -0.4308 | Yes |
| 46 | TNNT3 | NA | 38080 | -0.077 | -0.4229 | Yes |
| 47 | PDE4B | NA | 38334 | -0.078 | -0.4192 | Yes |
| 48 | CALM2 | NA | 38531 | -0.079 | -0.4143 | Yes |
| 49 | MYL2 | NA | 40305 | -0.090 | -0.4372 | Yes |
| 50 | MYH7 | NA | 41508 | -0.097 | -0.4488 | Yes |
| 51 | CALM3 | NA | 41720 | -0.099 | -0.4422 | Yes |
| 52 | SLC8A1 | NA | 41960 | -0.100 | -0.4358 | Yes |
| 53 | ACTN3 | NA | 42391 | -0.103 | -0.4327 | Yes |
| 54 | RNF207 | NA | 43008 | -0.107 | -0.4325 | Yes |
| 55 | TNNI1 | NA | 44982 | -0.121 | -0.4558 | Yes |
| 56 | TMEM38A | NA | 45403 | -0.124 | -0.4502 | Yes |
| 57 | CAV1 | NA | 45471 | -0.125 | -0.4381 | Yes |
| 58 | CASQ2 | NA | 45610 | -0.126 | -0.4271 | Yes |
| 59 | TNNI3K | NA | 47141 | -0.138 | -0.4404 | Yes |
| 60 | RYR2 | NA | 47165 | -0.138 | -0.4260 | Yes |
| 61 | TMEM38B | NA | 47270 | -0.139 | -0.4130 | Yes |
| 62 | KCNQ1 | NA | 47343 | -0.140 | -0.3994 | Yes |
| 63 | DMD | NA | 47602 | -0.142 | -0.3889 | Yes |
| 64 | C12orf57 | NA | 47740 | -0.143 | -0.3761 | Yes |
| 65 | DLG1 | NA | 47814 | -0.144 | -0.3620 | Yes |
| 66 | CASQ1 | NA | 48944 | -0.154 | -0.3662 | Yes |
| 67 | JUP | NA | 49496 | -0.160 | -0.3591 | Yes |
| 68 | PIK3CG | NA | 50070 | -0.166 | -0.3519 | Yes |
| 69 | CAV3 | NA | 50258 | -0.168 | -0.3374 | Yes |
| 70 | CLIC2 | NA | 50704 | -0.173 | -0.3270 | Yes |
| 71 | P2RX4 | NA | 50913 | -0.176 | -0.3120 | Yes |
| 72 | ADRA1B | NA | 50963 | -0.177 | -0.2940 | Yes |
| 73 | KCNJ2 | NA | 51031 | -0.177 | -0.2762 | Yes |
| 74 | KBTBD13 | NA | 51275 | -0.181 | -0.2613 | Yes |
| 75 | FKBP1B | NA | 51382 | -0.182 | -0.2437 | Yes |
| 76 | CACNA1C | NA | 51611 | -0.185 | -0.2281 | Yes |
| 77 | PLN | NA | 51923 | -0.190 | -0.2134 | Yes |
| 78 | SLC8A3 | NA | 52201 | -0.195 | -0.1976 | Yes |
| 79 | GRK2 | NA | 52225 | -0.195 | -0.1772 | Yes |
| 80 | GSTM2 | NA | 52778 | -0.206 | -0.1653 | Yes |
| 81 | AKAP9 | NA | 52953 | -0.210 | -0.1460 | Yes |
| 82 | TRPM4 | NA | 53090 | -0.213 | -0.1257 | Yes |
| 83 | DMPK | NA | 53110 | -0.214 | -0.1032 | Yes |
| 84 | RANGRF | NA | 53236 | -0.217 | -0.0823 | Yes |
| 85 | CTNNA3 | NA | 53643 | -0.229 | -0.0652 | Yes |
| 86 | MYL3 | NA | 54044 | -0.246 | -0.0462 | Yes |
| 87 | HRC | NA | 54236 | -0.256 | -0.0223 | Yes |
| 88 | ASB3 | NA | 54359 | -0.269 | 0.0043 | Yes |