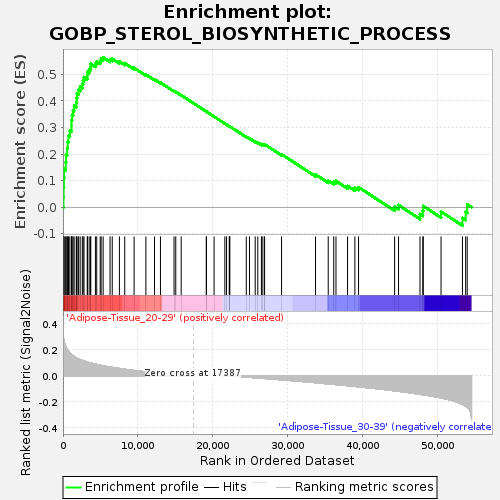
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_30-39_repos |
| Upregulated in class | Adipose-Tissue_20-29 |
| GeneSet | GOBP_STEROL_BIOSYNTHETIC_PROCESS |
| Enrichment Score (ES) | 0.5636242 |
| Normalized Enrichment Score (NES) | 1.556911 |
| Nominal p-value | 0.07317073 |
| FDR q-value | 0.4250575 |
| FWER p-Value | 0.922 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | NSDHL | NA | 60 | 0.295 | 0.0375 | Yes |
| 2 | LPCAT3 | NA | 74 | 0.289 | 0.0751 | Yes |
| 3 | ACAA2 | NA | 120 | 0.273 | 0.1099 | Yes |
| 4 | RAN | NA | 151 | 0.263 | 0.1437 | Yes |
| 5 | KPNB1 | NA | 366 | 0.226 | 0.1693 | Yes |
| 6 | CYP51A1 | NA | 407 | 0.219 | 0.1972 | Yes |
| 7 | FASN | NA | 549 | 0.204 | 0.2214 | Yes |
| 8 | NPC1L1 | NA | 625 | 0.198 | 0.2458 | Yes |
| 9 | G6PD | NA | 734 | 0.190 | 0.2686 | Yes |
| 10 | CYB5R2 | NA | 910 | 0.178 | 0.2887 | Yes |
| 11 | APOB | NA | 1127 | 0.164 | 0.3062 | Yes |
| 12 | HMGCS1 | NA | 1129 | 0.164 | 0.3276 | Yes |
| 13 | ACACB | NA | 1216 | 0.160 | 0.3470 | Yes |
| 14 | SQLE | NA | 1339 | 0.154 | 0.3649 | Yes |
| 15 | ACLY | NA | 1487 | 0.149 | 0.3817 | Yes |
| 16 | DHCR24 | NA | 1770 | 0.138 | 0.3946 | Yes |
| 17 | SC5D | NA | 1829 | 0.136 | 0.4114 | Yes |
| 18 | HMGCR | NA | 1874 | 0.135 | 0.4282 | Yes |
| 19 | ACACA | NA | 2079 | 0.129 | 0.4412 | Yes |
| 20 | DHCR7 | NA | 2292 | 0.124 | 0.4535 | Yes |
| 21 | INSIG1 | NA | 2576 | 0.118 | 0.4637 | Yes |
| 22 | SOD1 | NA | 2681 | 0.116 | 0.4770 | Yes |
| 23 | SREBF1 | NA | 2809 | 0.113 | 0.4895 | Yes |
| 24 | ACAT2 | NA | 3251 | 0.106 | 0.4952 | Yes |
| 25 | MSMO1 | NA | 3258 | 0.105 | 0.5088 | Yes |
| 26 | EBP | NA | 3491 | 0.102 | 0.5179 | Yes |
| 27 | SEC14L2 | NA | 3662 | 0.099 | 0.5277 | Yes |
| 28 | ELOVL6 | NA | 3714 | 0.098 | 0.5396 | Yes |
| 29 | FDFT1 | NA | 4332 | 0.089 | 0.5399 | Yes |
| 30 | SCD | NA | 4505 | 0.087 | 0.5482 | Yes |
| 31 | SCAP | NA | 4966 | 0.081 | 0.5504 | Yes |
| 32 | APOE | NA | 5094 | 0.080 | 0.5585 | Yes |
| 33 | FDPS | NA | 5363 | 0.077 | 0.5636 | Yes |
| 34 | APOA4 | NA | 6288 | 0.067 | 0.5554 | No |
| 35 | MVK | NA | 6579 | 0.064 | 0.5585 | No |
| 36 | HSD17B7 | NA | 7536 | 0.056 | 0.5482 | No |
| 37 | MIR548P | NA | 8246 | 0.050 | 0.5417 | No |
| 38 | GPAM | NA | 9499 | 0.040 | 0.5241 | No |
| 39 | HMGCS2 | NA | 11076 | 0.030 | 0.4990 | No |
| 40 | IDI1 | NA | 12228 | 0.023 | 0.4809 | No |
| 41 | SREBF2 | NA | 13022 | 0.019 | 0.4688 | No |
| 42 | CH25H | NA | 14838 | 0.010 | 0.4369 | No |
| 43 | MVD | NA | 15067 | 0.009 | 0.4339 | No |
| 44 | CFTR | NA | 15792 | 0.006 | 0.4214 | No |
| 45 | MIR30C1 | NA | 19152 | 0.000 | 0.3598 | No |
| 46 | MIR182 | NA | 19158 | 0.000 | 0.3597 | No |
| 47 | MIR185 | NA | 20187 | 0.000 | 0.3408 | No |
| 48 | PLPP6 | NA | 21626 | -0.001 | 0.3146 | No |
| 49 | PMVK | NA | 21849 | -0.002 | 0.3108 | No |
| 50 | TM7SF2 | NA | 22211 | -0.003 | 0.3046 | No |
| 51 | FGF1 | NA | 22269 | -0.003 | 0.3040 | No |
| 52 | MBTPS2 | NA | 22274 | -0.003 | 0.3043 | No |
| 53 | APOA1 | NA | 24488 | -0.011 | 0.2652 | No |
| 54 | IDI2 | NA | 24918 | -0.012 | 0.2589 | No |
| 55 | MIR96 | NA | 25681 | -0.015 | 0.2470 | No |
| 56 | NFYA | NA | 26028 | -0.017 | 0.2428 | No |
| 57 | CYB5R3 | NA | 26510 | -0.019 | 0.2365 | No |
| 58 | ABCG1 | NA | 26607 | -0.019 | 0.2373 | No |
| 59 | ERLIN2 | NA | 26879 | -0.021 | 0.2350 | No |
| 60 | ERLIN1 | NA | 26937 | -0.021 | 0.2367 | No |
| 61 | LSS | NA | 29202 | -0.031 | 0.1993 | No |
| 62 | PRKAG2 | NA | 33741 | -0.053 | 0.1230 | No |
| 63 | MBTPS1 | NA | 35438 | -0.063 | 0.1000 | No |
| 64 | CYP7A1 | NA | 36172 | -0.066 | 0.0953 | No |
| 65 | ARV1 | NA | 36474 | -0.068 | 0.0987 | No |
| 66 | APOA5 | NA | 38020 | -0.076 | 0.0803 | No |
| 67 | LBR | NA | 38998 | -0.082 | 0.0731 | No |
| 68 | GGPS1 | NA | 39488 | -0.085 | 0.0753 | No |
| 69 | CES1 | NA | 44307 | -0.116 | 0.0020 | No |
| 70 | INSIG2 | NA | 44832 | -0.120 | 0.0081 | No |
| 71 | PRKAA1 | NA | 47699 | -0.143 | -0.0258 | No |
| 72 | NFYC | NA | 48073 | -0.147 | -0.0135 | No |
| 73 | PRKAA2 | NA | 48137 | -0.147 | 0.0046 | No |
| 74 | SP1 | NA | 50511 | -0.171 | -0.0166 | No |
| 75 | CYB5R1 | NA | 53373 | -0.220 | -0.0403 | No |
| 76 | NFYB | NA | 53802 | -0.235 | -0.0174 | No |
| 77 | FAXDC2 | NA | 54008 | -0.244 | 0.0107 | No |