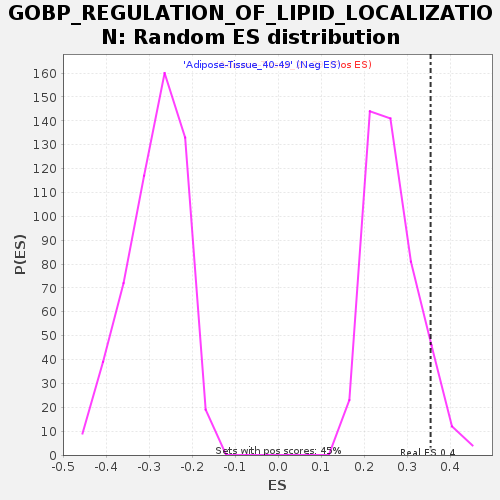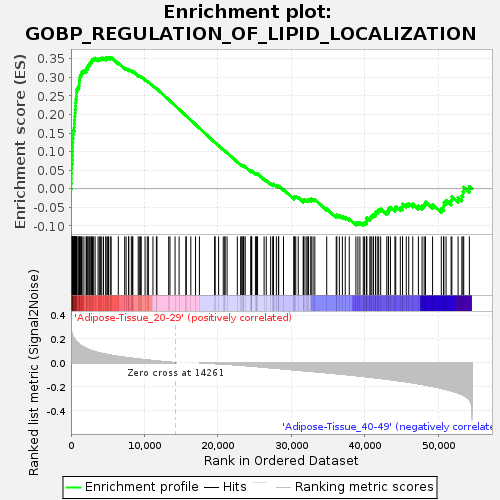
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Adipose-Tissue.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_40-49.Adipose-Tissue.cls #Adipose-Tissue_20-29_versus_Adipose-Tissue_40-49_repos |
| Phenotype | Adipose-Tissue.cls#Adipose-Tissue_20-29_versus_Adipose-Tissue_40-49_repos |
| Upregulated in class | Adipose-Tissue_20-29 |
| GeneSet | GOBP_REGULATION_OF_LIPID_LOCALIZATION |
| Enrichment Score (ES) | 0.353791 |
| Normalized Enrichment Score (NES) | 1.3395046 |
| Nominal p-value | 0.079822615 |
| FDR q-value | 0.7614185 |
| FWER p-Value | 0.989 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ACSL1 | NA | 26 | 0.289 | 0.0146 | Yes |
| 2 | FURIN | NA | 54 | 0.267 | 0.0281 | Yes |
| 3 | DBI | NA | 57 | 0.266 | 0.0419 | Yes |
| 4 | PPARG | NA | 81 | 0.255 | 0.0548 | Yes |
| 5 | TSPO | NA | 119 | 0.246 | 0.0670 | Yes |
| 6 | PRELID1 | NA | 183 | 0.232 | 0.0779 | Yes |
| 7 | PNPLA2 | NA | 195 | 0.230 | 0.0897 | Yes |
| 8 | EHD1 | NA | 208 | 0.228 | 0.1014 | Yes |
| 9 | ACACB | NA | 217 | 0.226 | 0.1131 | Yes |
| 10 | AKT1 | NA | 226 | 0.224 | 0.1247 | Yes |
| 11 | ABHD5 | NA | 246 | 0.222 | 0.1359 | Yes |
| 12 | MIR27A | NA | 301 | 0.216 | 0.1462 | Yes |
| 13 | C1QTNF1 | NA | 316 | 0.214 | 0.1571 | Yes |
| 14 | IL6 | NA | 430 | 0.201 | 0.1655 | Yes |
| 15 | CIDEA | NA | 445 | 0.200 | 0.1757 | Yes |
| 16 | ZC3H12A | NA | 476 | 0.197 | 0.1855 | Yes |
| 17 | APOB | NA | 504 | 0.194 | 0.1951 | Yes |
| 18 | FITM2 | NA | 548 | 0.191 | 0.2043 | Yes |
| 19 | LPCAT3 | NA | 560 | 0.190 | 0.2140 | Yes |
| 20 | PPARD | NA | 604 | 0.187 | 0.2230 | Yes |
| 21 | THBS1 | NA | 623 | 0.185 | 0.2324 | Yes |
| 22 | ADIPOQ | NA | 659 | 0.184 | 0.2413 | Yes |
| 23 | CCK | NA | 743 | 0.177 | 0.2490 | Yes |
| 24 | PLIN5 | NA | 750 | 0.177 | 0.2582 | Yes |
| 25 | AVPR1A | NA | 761 | 0.176 | 0.2672 | Yes |
| 26 | LPL | NA | 929 | 0.166 | 0.2728 | Yes |
| 27 | PRKCD | NA | 1070 | 0.158 | 0.2785 | Yes |
| 28 | PPARA | NA | 1116 | 0.155 | 0.2857 | Yes |
| 29 | NTSR1 | NA | 1133 | 0.155 | 0.2935 | Yes |
| 30 | LIPG | NA | 1194 | 0.152 | 0.3003 | Yes |
| 31 | POMC | NA | 1304 | 0.145 | 0.3059 | Yes |
| 32 | NFKB1 | NA | 1437 | 0.140 | 0.3108 | Yes |
| 33 | SCP2D1 | NA | 1512 | 0.137 | 0.3166 | Yes |
| 34 | NFKBIA | NA | 1755 | 0.128 | 0.3188 | Yes |
| 35 | ERFE | NA | 2056 | 0.119 | 0.3196 | Yes |
| 36 | ABCA1 | NA | 2102 | 0.118 | 0.3249 | Yes |
| 37 | LAMTOR1 | NA | 2284 | 0.113 | 0.3275 | Yes |
| 38 | PLA2G10 | NA | 2322 | 0.112 | 0.3326 | Yes |
| 39 | ADORA1 | NA | 2486 | 0.108 | 0.3352 | Yes |
| 40 | MAP2K6 | NA | 2590 | 0.106 | 0.3389 | Yes |
| 41 | IRS2 | NA | 2719 | 0.103 | 0.3419 | Yes |
| 42 | CRY1 | NA | 2804 | 0.101 | 0.3456 | Yes |
| 43 | IL1B | NA | 2922 | 0.099 | 0.3486 | Yes |
| 44 | TAC1 | NA | 3085 | 0.096 | 0.3507 | Yes |
| 45 | ATP8A2 | NA | 3285 | 0.092 | 0.3518 | Yes |
| 46 | ITGB3 | NA | 3685 | 0.085 | 0.3489 | Yes |
| 47 | TRIAP1 | NA | 3857 | 0.083 | 0.3501 | Yes |
| 48 | NKX3-1 | NA | 4034 | 0.080 | 0.3511 | Yes |
| 49 | AKT2 | NA | 4189 | 0.078 | 0.3523 | Yes |
| 50 | ANXA2 | NA | 4427 | 0.075 | 0.3518 | Yes |
| 51 | SREBF2 | NA | 4736 | 0.071 | 0.3499 | Yes |
| 52 | ABCG8 | NA | 4774 | 0.070 | 0.3528 | Yes |
| 53 | OSBPL11 | NA | 4992 | 0.068 | 0.3524 | Yes |
| 54 | PER2 | NA | 5105 | 0.066 | 0.3538 | Yes |
| 55 | PLIN3 | NA | 5342 | 0.064 | 0.3528 | No |
| 56 | OXT | NA | 5467 | 0.062 | 0.3537 | No |
| 57 | PLIN2 | NA | 6440 | 0.052 | 0.3386 | No |
| 58 | ATP1A2 | NA | 7309 | 0.045 | 0.3250 | No |
| 59 | AVPR1B | NA | 7504 | 0.043 | 0.3237 | No |
| 60 | ARL6IP1 | NA | 7837 | 0.041 | 0.3197 | No |
| 61 | AVP | NA | 7863 | 0.040 | 0.3214 | No |
| 62 | FIS1 | NA | 8211 | 0.038 | 0.3169 | No |
| 63 | APOE | NA | 8346 | 0.037 | 0.3164 | No |
| 64 | TNF | NA | 8420 | 0.036 | 0.3169 | No |
| 65 | PTPN2 | NA | 9151 | 0.031 | 0.3051 | No |
| 66 | FABP3 | NA | 9296 | 0.030 | 0.3040 | No |
| 67 | ITGB1 | NA | 9477 | 0.029 | 0.3022 | No |
| 68 | AGT | NA | 9553 | 0.028 | 0.3023 | No |
| 69 | TRH | NA | 10083 | 0.024 | 0.2939 | No |
| 70 | VSTM2A | NA | 10385 | 0.023 | 0.2895 | No |
| 71 | RETN | NA | 10536 | 0.022 | 0.2879 | No |
| 72 | PLA2G4A | NA | 11160 | 0.017 | 0.2773 | No |
| 73 | ARL6IP5 | NA | 11624 | 0.015 | 0.2696 | No |
| 74 | MIR10B | NA | 11703 | 0.014 | 0.2689 | No |
| 75 | MIR27B | NA | 13300 | 0.005 | 0.2398 | No |
| 76 | PTPN11 | NA | 13472 | 0.004 | 0.2369 | No |
| 77 | CD36 | NA | 14167 | 0.001 | 0.2242 | No |
| 78 | MIR33A | NA | 14718 | 0.000 | 0.2141 | No |
| 79 | MIR128-1 | NA | 15637 | 0.000 | 0.1972 | No |
| 80 | MIR30C1 | NA | 15693 | 0.000 | 0.1962 | No |
| 81 | MIR301B | NA | 16314 | 0.000 | 0.1848 | No |
| 82 | MIR302A | NA | 16954 | 0.000 | 0.1730 | No |
| 83 | MIR148A | NA | 17484 | 0.000 | 0.1633 | No |
| 84 | HRH3 | NA | 19553 | -0.005 | 0.1256 | No |
| 85 | SIRT1 | NA | 19629 | -0.006 | 0.1245 | No |
| 86 | TNFSF11 | NA | 20099 | -0.007 | 0.1163 | No |
| 87 | NUS1 | NA | 20729 | -0.009 | 0.1052 | No |
| 88 | ABCA2 | NA | 20952 | -0.010 | 0.1016 | No |
| 89 | MIR130B | NA | 20966 | -0.010 | 0.1019 | No |
| 90 | GALR1 | NA | 21251 | -0.011 | 0.0973 | No |
| 91 | ASXL1 | NA | 22643 | -0.017 | 0.0726 | No |
| 92 | AGTR1 | NA | 23095 | -0.019 | 0.0653 | No |
| 93 | CYP19A1 | NA | 23243 | -0.020 | 0.0637 | No |
| 94 | CRH | NA | 23384 | -0.021 | 0.0622 | No |
| 95 | ENPP7 | NA | 23471 | -0.021 | 0.0617 | No |
| 96 | MIR33B | NA | 23531 | -0.021 | 0.0617 | No |
| 97 | CAV1 | NA | 23720 | -0.022 | 0.0594 | No |
| 98 | APOC1 | NA | 24485 | -0.026 | 0.0467 | No |
| 99 | KMO | NA | 24514 | -0.026 | 0.0476 | No |
| 100 | ABCA3 | NA | 24577 | -0.027 | 0.0478 | No |
| 101 | MIR26A1 | NA | 24614 | -0.027 | 0.0486 | No |
| 102 | APOA5 | NA | 25110 | -0.029 | 0.0410 | No |
| 103 | ABCG5 | NA | 25279 | -0.030 | 0.0395 | No |
| 104 | EDN1 | NA | 25293 | -0.030 | 0.0408 | No |
| 105 | ITGAV | NA | 25398 | -0.031 | 0.0405 | No |
| 106 | APOA4 | NA | 26291 | -0.036 | 0.0260 | No |
| 107 | LRP1 | NA | 26584 | -0.037 | 0.0226 | No |
| 108 | NR1H2 | NA | 27180 | -0.040 | 0.0138 | No |
| 109 | NAXE | NA | 27486 | -0.042 | 0.0103 | No |
| 110 | PLA2G3 | NA | 27512 | -0.042 | 0.0121 | No |
| 111 | PLTP | NA | 27569 | -0.043 | 0.0133 | No |
| 112 | ACSL5 | NA | 27960 | -0.045 | 0.0085 | No |
| 113 | HILPDA | NA | 28241 | -0.047 | 0.0057 | No |
| 114 | CES1 | NA | 28278 | -0.047 | 0.0075 | No |
| 115 | LEP | NA | 28931 | -0.050 | -0.0018 | No |
| 116 | SPP1 | NA | 30354 | -0.058 | -0.0250 | No |
| 117 | GAL | NA | 30361 | -0.058 | -0.0220 | No |
| 118 | APOC4 | NA | 30456 | -0.058 | -0.0207 | No |
| 119 | PCSK9 | NA | 30599 | -0.059 | -0.0202 | No |
| 120 | AGTR2 | NA | 30940 | -0.061 | -0.0233 | No |
| 121 | APOA2 | NA | 31625 | -0.065 | -0.0325 | No |
| 122 | TNFRSF11A | NA | 31638 | -0.065 | -0.0293 | No |
| 123 | PTGES | NA | 31854 | -0.066 | -0.0298 | No |
| 124 | CYP4F2 | NA | 32108 | -0.068 | -0.0309 | No |
| 125 | BMP6 | NA | 32252 | -0.068 | -0.0300 | No |
| 126 | MSR1 | NA | 32348 | -0.069 | -0.0281 | No |
| 127 | APOA1 | NA | 32659 | -0.071 | -0.0301 | No |
| 128 | MIR758 | NA | 32692 | -0.071 | -0.0270 | No |
| 129 | SCP2 | NA | 32956 | -0.072 | -0.0281 | No |
| 130 | APOC3 | NA | 33196 | -0.073 | -0.0286 | No |
| 131 | PSEN1 | NA | 34816 | -0.082 | -0.0541 | No |
| 132 | REN | NA | 36110 | -0.090 | -0.0732 | No |
| 133 | GDF9 | NA | 36197 | -0.090 | -0.0700 | No |
| 134 | CRP | NA | 36539 | -0.092 | -0.0715 | No |
| 135 | SYK | NA | 36951 | -0.095 | -0.0741 | No |
| 136 | SSTR4 | NA | 37339 | -0.097 | -0.0761 | No |
| 137 | ABCB4 | NA | 37883 | -0.101 | -0.0808 | No |
| 138 | MYB | NA | 38789 | -0.106 | -0.0919 | No |
| 139 | OSBPL6 | NA | 39064 | -0.108 | -0.0913 | No |
| 140 | SHH | NA | 39337 | -0.110 | -0.0906 | No |
| 141 | NR1H3 | NA | 39796 | -0.113 | -0.0931 | No |
| 142 | APOC2 | NA | 39972 | -0.114 | -0.0904 | No |
| 143 | GPS2 | NA | 40214 | -0.116 | -0.0888 | No |
| 144 | C3 | NA | 40234 | -0.116 | -0.0831 | No |
| 145 | PON1 | NA | 40285 | -0.116 | -0.0779 | No |
| 146 | ZDHHC8 | NA | 40689 | -0.119 | -0.0791 | No |
| 147 | EEPD1 | NA | 40826 | -0.120 | -0.0753 | No |
| 148 | STXBP1 | NA | 40965 | -0.121 | -0.0715 | No |
| 149 | NPY5R | NA | 41182 | -0.122 | -0.0691 | No |
| 150 | DISP3 | NA | 41453 | -0.124 | -0.0676 | No |
| 151 | ABCA7 | NA | 41494 | -0.125 | -0.0618 | No |
| 152 | SLC12A2 | NA | 41768 | -0.127 | -0.0602 | No |
| 153 | ADORA2A | NA | 41895 | -0.128 | -0.0558 | No |
| 154 | FITM1 | NA | 42164 | -0.130 | -0.0540 | No |
| 155 | GHRL | NA | 42994 | -0.136 | -0.0621 | No |
| 156 | EGF | NA | 43204 | -0.138 | -0.0587 | No |
| 157 | ARV1 | NA | 43231 | -0.138 | -0.0520 | No |
| 158 | ASXL2 | NA | 43494 | -0.140 | -0.0495 | No |
| 159 | CRHR1 | NA | 44103 | -0.145 | -0.0531 | No |
| 160 | OSBPL8 | NA | 44215 | -0.146 | -0.0475 | No |
| 161 | GRM7 | NA | 44837 | -0.151 | -0.0511 | No |
| 162 | KDM5B | NA | 45138 | -0.153 | -0.0486 | No |
| 163 | ABCA12 | NA | 45141 | -0.153 | -0.0406 | No |
| 164 | P2RX4 | NA | 45648 | -0.158 | -0.0417 | No |
| 165 | ABCG1 | NA | 45981 | -0.161 | -0.0394 | No |
| 166 | CYP4A11 | NA | 46532 | -0.166 | -0.0408 | No |
| 167 | RAB3GAP1 | NA | 47301 | -0.174 | -0.0458 | No |
| 168 | IKBKE | NA | 47799 | -0.180 | -0.0456 | No |
| 169 | FASLG | NA | 48084 | -0.183 | -0.0412 | No |
| 170 | LDLRAP1 | NA | 48265 | -0.185 | -0.0349 | No |
| 171 | LRAT | NA | 49230 | -0.196 | -0.0424 | No |
| 172 | TMF1 | NA | 50418 | -0.212 | -0.0531 | No |
| 173 | ABCA8 | NA | 50723 | -0.216 | -0.0474 | No |
| 174 | CYP8B1 | NA | 50766 | -0.217 | -0.0369 | No |
| 175 | PTCH1 | NA | 51067 | -0.221 | -0.0309 | No |
| 176 | ATP8A1 | NA | 51750 | -0.232 | -0.0313 | No |
| 177 | CETP | NA | 51855 | -0.234 | -0.0209 | No |
| 178 | SCARB1 | NA | 52704 | -0.251 | -0.0234 | No |
| 179 | TMEM30A | NA | 53213 | -0.264 | -0.0190 | No |
| 180 | YJEFN3 | NA | 53361 | -0.268 | -0.0076 | No |
| 181 | PLA2R1 | NA | 53470 | -0.272 | 0.0046 | No |
| 182 | TTC39B | NA | 54238 | -0.307 | 0.0065 | No |