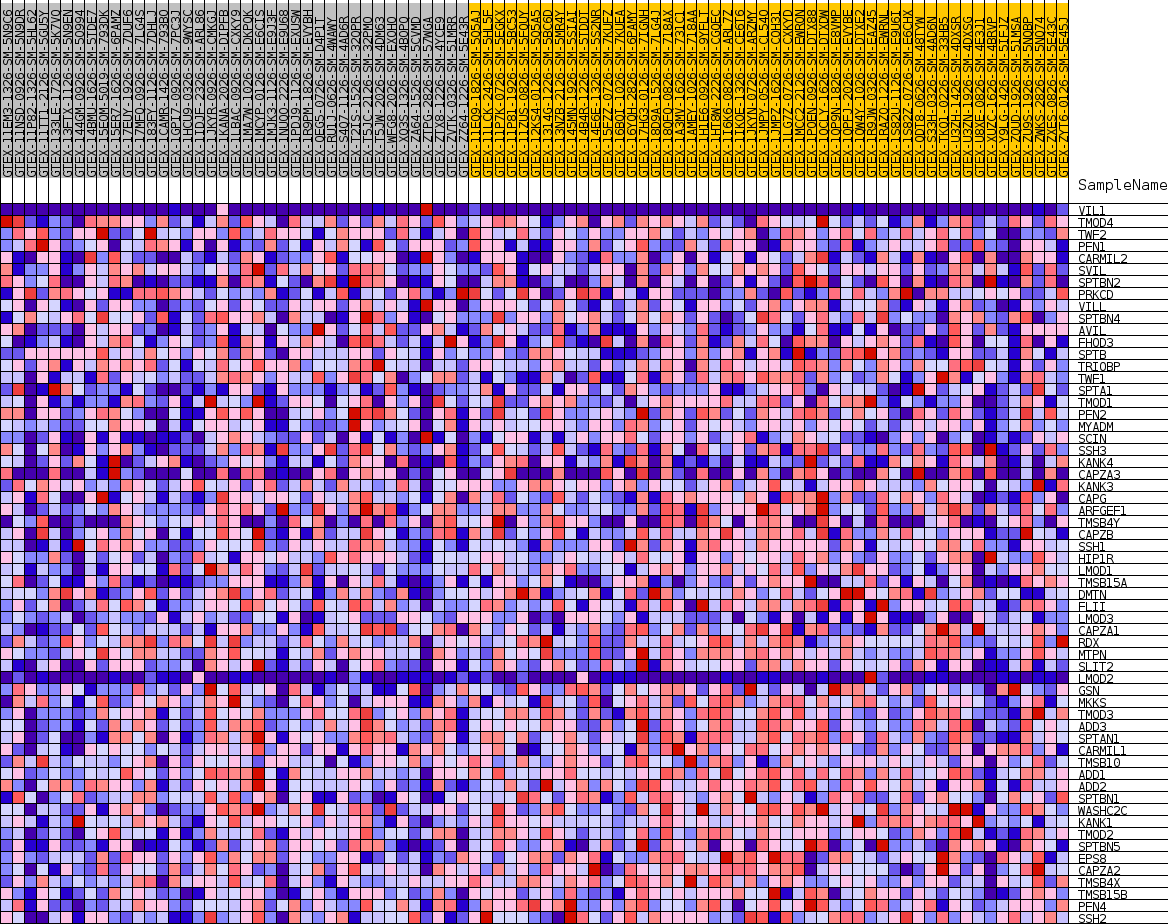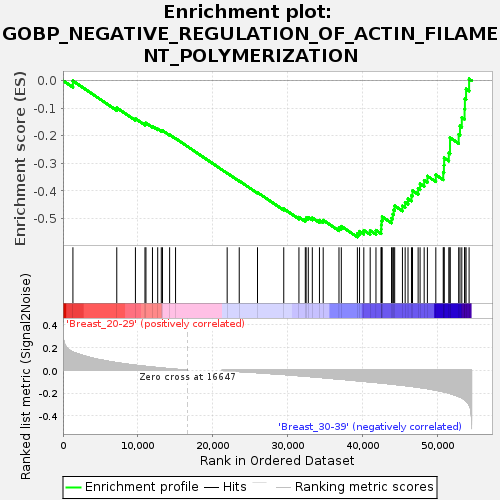
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_20-29_versus_Breast_30-39.Breast.cls #Breast_20-29_versus_Breast_30-39_repos |
| Phenotype | Breast.cls#Breast_20-29_versus_Breast_30-39_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_NEGATIVE_REGULATION_OF_ACTIN_FILAMENT_POLYMERIZATION |
| Enrichment Score (ES) | -0.567469 |
| Normalized Enrichment Score (NES) | -1.7166724 |
| Nominal p-value | 0.0076190475 |
| FDR q-value | 0.93206817 |
| FWER p-Value | 0.667 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | VIL1 | NA | 1325 | 0.159 | -0.0011 | No |
| 2 | TMOD4 | NA | 7184 | 0.067 | -0.0988 | No |
| 3 | TWF2 | NA | 9677 | 0.044 | -0.1381 | No |
| 4 | PFN1 | NA | 10952 | 0.035 | -0.1564 | No |
| 5 | CARMIL2 | NA | 11083 | 0.034 | -0.1538 | No |
| 6 | SVIL | NA | 11954 | 0.028 | -0.1657 | No |
| 7 | SPTBN2 | NA | 12653 | 0.023 | -0.1751 | No |
| 8 | PRKCD | NA | 13133 | 0.020 | -0.1810 | No |
| 9 | VILL | NA | 13291 | 0.019 | -0.1810 | No |
| 10 | SPTBN4 | NA | 14247 | 0.014 | -0.1966 | No |
| 11 | AVIL | NA | 15038 | 0.009 | -0.2097 | No |
| 12 | FHOD3 | NA | 21935 | -0.004 | -0.3356 | No |
| 13 | SPTB | NA | 23547 | -0.010 | -0.3637 | No |
| 14 | TRIOBP | NA | 25988 | -0.019 | -0.4056 | No |
| 15 | TWF1 | NA | 29495 | -0.035 | -0.4648 | No |
| 16 | SPTA1 | NA | 31524 | -0.046 | -0.4953 | No |
| 17 | TMOD1 | NA | 32375 | -0.050 | -0.5036 | No |
| 18 | PFN2 | NA | 32484 | -0.051 | -0.4981 | No |
| 19 | MYADM | NA | 32775 | -0.052 | -0.4958 | No |
| 20 | SCIN | NA | 33312 | -0.056 | -0.4975 | No |
| 21 | SSH3 | NA | 34266 | -0.061 | -0.5062 | No |
| 22 | KANK4 | NA | 34766 | -0.064 | -0.5061 | No |
| 23 | CAPZA3 | NA | 36874 | -0.076 | -0.5337 | No |
| 24 | KANK3 | NA | 37187 | -0.077 | -0.5281 | No |
| 25 | CAPG | NA | 39333 | -0.090 | -0.5543 | Yes |
| 26 | ARFGEF1 | NA | 39621 | -0.092 | -0.5462 | Yes |
| 27 | TMSB4Y | NA | 40192 | -0.095 | -0.5428 | Yes |
| 28 | CAPZB | NA | 41045 | -0.100 | -0.5438 | Yes |
| 29 | SSH1 | NA | 41816 | -0.105 | -0.5426 | Yes |
| 30 | HIP1R | NA | 42512 | -0.110 | -0.5393 | Yes |
| 31 | LMOD1 | NA | 42521 | -0.110 | -0.5235 | Yes |
| 32 | TMSB15A | NA | 42565 | -0.110 | -0.5083 | Yes |
| 33 | DMTN | NA | 42643 | -0.111 | -0.4936 | Yes |
| 34 | FLII | NA | 43896 | -0.119 | -0.4992 | Yes |
| 35 | LMOD3 | NA | 44055 | -0.120 | -0.4845 | Yes |
| 36 | CAPZA1 | NA | 44180 | -0.121 | -0.4692 | Yes |
| 37 | RDX | NA | 44318 | -0.122 | -0.4539 | Yes |
| 38 | MTPN | NA | 45354 | -0.130 | -0.4540 | Yes |
| 39 | SLIT2 | NA | 45718 | -0.132 | -0.4414 | Yes |
| 40 | LMOD2 | NA | 46098 | -0.135 | -0.4286 | Yes |
| 41 | GSN | NA | 46561 | -0.139 | -0.4168 | Yes |
| 42 | MKKS | NA | 46699 | -0.140 | -0.3988 | Yes |
| 43 | TMOD3 | NA | 47450 | -0.147 | -0.3911 | Yes |
| 44 | ADD3 | NA | 47712 | -0.150 | -0.3741 | Yes |
| 45 | SPTAN1 | NA | 48258 | -0.155 | -0.3616 | Yes |
| 46 | CARMIL1 | NA | 48688 | -0.160 | -0.3462 | Yes |
| 47 | TMSB10 | NA | 49814 | -0.173 | -0.3417 | Yes |
| 48 | ADD1 | NA | 50805 | -0.186 | -0.3327 | Yes |
| 49 | ADD2 | NA | 50897 | -0.188 | -0.3071 | Yes |
| 50 | SPTBN1 | NA | 50925 | -0.188 | -0.2801 | Yes |
| 51 | WASHC2C | NA | 51550 | -0.198 | -0.2627 | Yes |
| 52 | KANK1 | NA | 51712 | -0.201 | -0.2363 | Yes |
| 53 | TMOD2 | NA | 51713 | -0.201 | -0.2070 | Yes |
| 54 | SPTBN5 | NA | 52869 | -0.228 | -0.1950 | Yes |
| 55 | EPS8 | NA | 53042 | -0.233 | -0.1642 | Yes |
| 56 | CAPZA2 | NA | 53287 | -0.241 | -0.1336 | Yes |
| 57 | TMSB4X | NA | 53656 | -0.255 | -0.1032 | Yes |
| 58 | TMSB15B | NA | 53699 | -0.257 | -0.0664 | Yes |
| 59 | PFN4 | NA | 53855 | -0.267 | -0.0304 | Yes |
| 60 | SSH2 | NA | 54260 | -0.301 | 0.0061 | Yes |