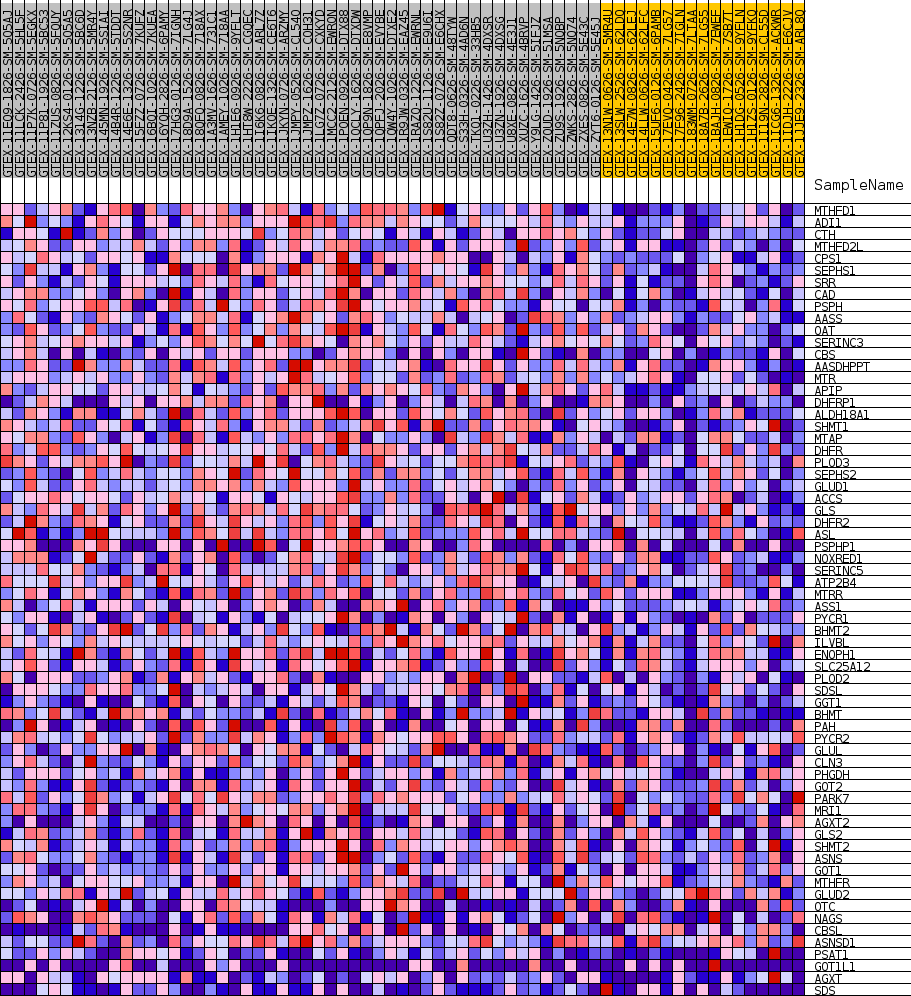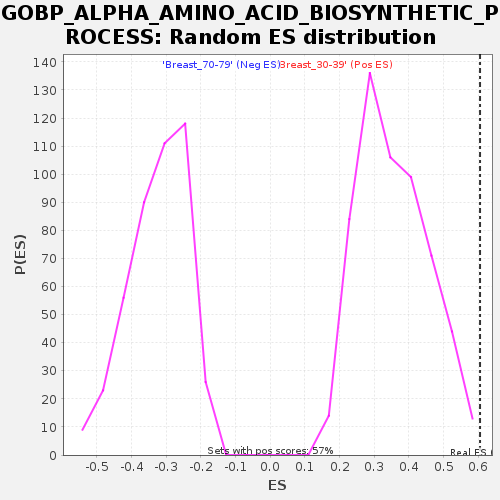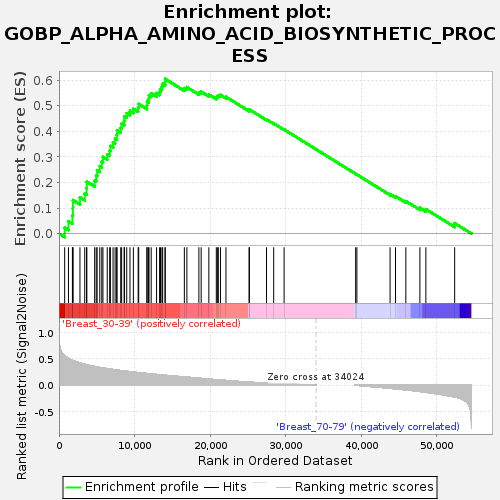
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_ALPHA_AMINO_ACID_BIOSYNTHETIC_PROCESS |
| Enrichment Score (ES) | 0.6052333 |
| Normalized Enrichment Score (NES) | 1.7056729 |
| Nominal p-value | 0.0035273368 |
| FDR q-value | 0.3467245 |
| FWER p-Value | 0.633 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | MTHFD1 | NA | 751 | 0.557 | 0.0234 | Yes |
| 2 | ADI1 | NA | 1253 | 0.507 | 0.0481 | Yes |
| 3 | CTH | NA | 1775 | 0.469 | 0.0699 | Yes |
| 4 | MTHFD2L | NA | 1836 | 0.466 | 0.0999 | Yes |
| 5 | CPS1 | NA | 1847 | 0.465 | 0.1308 | Yes |
| 6 | SEPHS1 | NA | 2774 | 0.419 | 0.1418 | Yes |
| 7 | SRR | NA | 3405 | 0.394 | 0.1566 | Yes |
| 8 | CAD | NA | 3652 | 0.386 | 0.1779 | Yes |
| 9 | PSPH | NA | 3679 | 0.384 | 0.2031 | Yes |
| 10 | AASS | NA | 4720 | 0.353 | 0.2076 | Yes |
| 11 | OAT | NA | 4925 | 0.347 | 0.2270 | Yes |
| 12 | SERINC3 | NA | 5046 | 0.344 | 0.2478 | Yes |
| 13 | CBS | NA | 5399 | 0.334 | 0.2637 | Yes |
| 14 | AASDHPPT | NA | 5663 | 0.328 | 0.2808 | Yes |
| 15 | MTR | NA | 5816 | 0.325 | 0.2997 | Yes |
| 16 | APIP | NA | 6386 | 0.312 | 0.3101 | Yes |
| 17 | DHFRP1 | NA | 6709 | 0.305 | 0.3246 | Yes |
| 18 | ALDH18A1 | NA | 6807 | 0.303 | 0.3430 | Yes |
| 19 | SHMT1 | NA | 7155 | 0.296 | 0.3564 | Yes |
| 20 | MTAP | NA | 7426 | 0.290 | 0.3709 | Yes |
| 21 | DHFR | NA | 7628 | 0.286 | 0.3863 | Yes |
| 22 | PLOD3 | NA | 7684 | 0.285 | 0.4044 | Yes |
| 23 | SEPHS2 | NA | 8171 | 0.276 | 0.4139 | Yes |
| 24 | GLUD1 | NA | 8290 | 0.274 | 0.4300 | Yes |
| 25 | ACCS | NA | 8623 | 0.268 | 0.4418 | Yes |
| 26 | GLS | NA | 8664 | 0.267 | 0.4590 | Yes |
| 27 | DHFR2 | NA | 8964 | 0.262 | 0.4710 | Yes |
| 28 | ASL | NA | 9394 | 0.254 | 0.4801 | Yes |
| 29 | PSPHP1 | NA | 9846 | 0.247 | 0.4883 | Yes |
| 30 | NOXRED1 | NA | 10479 | 0.237 | 0.4925 | Yes |
| 31 | SERINC5 | NA | 10552 | 0.236 | 0.5070 | Yes |
| 32 | ATP2B4 | NA | 11629 | 0.220 | 0.5019 | Yes |
| 33 | MTRR | NA | 11668 | 0.219 | 0.5158 | Yes |
| 34 | ASS1 | NA | 11856 | 0.216 | 0.5269 | Yes |
| 35 | PYCR1 | NA | 11910 | 0.216 | 0.5403 | Yes |
| 36 | BHMT2 | NA | 12202 | 0.212 | 0.5491 | Yes |
| 37 | ILVBL | NA | 12916 | 0.202 | 0.5495 | Yes |
| 38 | ENOPH1 | NA | 13314 | 0.196 | 0.5554 | Yes |
| 39 | SLC25A12 | NA | 13434 | 0.194 | 0.5662 | Yes |
| 40 | PLOD2 | NA | 13626 | 0.192 | 0.5755 | Yes |
| 41 | SDSL | NA | 13723 | 0.190 | 0.5864 | Yes |
| 42 | GGT1 | NA | 14045 | 0.186 | 0.5930 | Yes |
| 43 | BHMT | NA | 14055 | 0.186 | 0.6052 | Yes |
| 44 | PAH | NA | 16594 | 0.154 | 0.5690 | No |
| 45 | PYCR2 | NA | 16931 | 0.150 | 0.5728 | No |
| 46 | GLUL | NA | 18517 | 0.130 | 0.5525 | No |
| 47 | CLN3 | NA | 18819 | 0.126 | 0.5554 | No |
| 48 | PHGDH | NA | 19837 | 0.113 | 0.5443 | No |
| 49 | GOT2 | NA | 20824 | 0.102 | 0.5330 | No |
| 50 | PARK7 | NA | 20966 | 0.100 | 0.5371 | No |
| 51 | MRI1 | NA | 21090 | 0.098 | 0.5414 | No |
| 52 | AGXT2 | NA | 21376 | 0.095 | 0.5425 | No |
| 53 | GLS2 | NA | 22103 | 0.087 | 0.5350 | No |
| 54 | SHMT2 | NA | 25168 | 0.056 | 0.4825 | No |
| 55 | ASNS | NA | 25196 | 0.055 | 0.4858 | No |
| 56 | GOT1 | NA | 27470 | 0.036 | 0.4465 | No |
| 57 | MTHFR | NA | 28420 | 0.030 | 0.4311 | No |
| 58 | GLUD2 | NA | 29800 | 0.021 | 0.4072 | No |
| 59 | OTC | NA | 39269 | -0.002 | 0.2337 | No |
| 60 | NAGS | NA | 39434 | -0.004 | 0.2310 | No |
| 61 | CBSL | NA | 43828 | -0.060 | 0.1544 | No |
| 62 | ASNSD1 | NA | 44545 | -0.070 | 0.1459 | No |
| 63 | PSAT1 | NA | 45914 | -0.090 | 0.1269 | No |
| 64 | GOT1L1 | NA | 47773 | -0.121 | 0.1009 | No |
| 65 | AGXT | NA | 48565 | -0.136 | 0.0955 | No |
| 66 | SDS | NA | 52377 | -0.225 | 0.0406 | No |