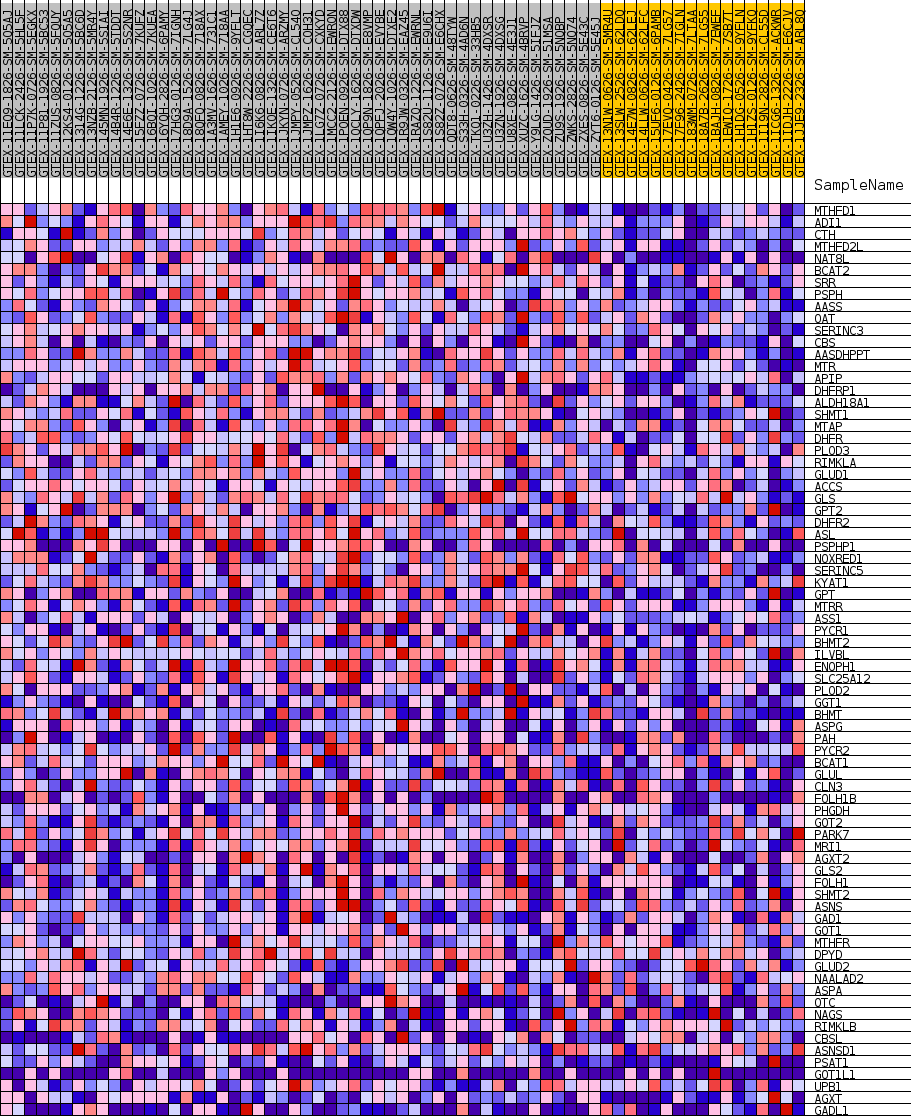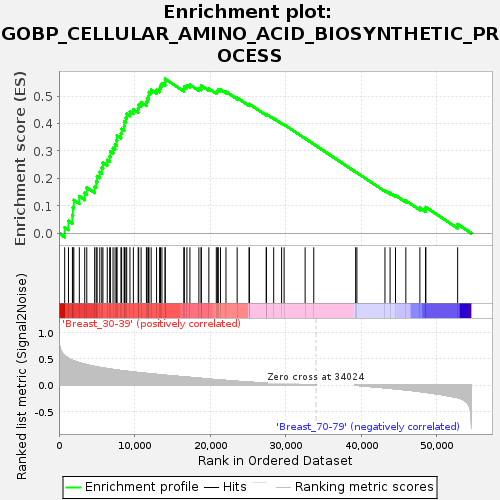
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_CELLULAR_AMINO_ACID_BIOSYNTHETIC_PROCESS |
| Enrichment Score (ES) | 0.56225115 |
| Normalized Enrichment Score (NES) | 1.6745325 |
| Nominal p-value | 0.0017271157 |
| FDR q-value | 0.2800045 |
| FWER p-Value | 0.737 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | MTHFD1 | NA | 751 | 0.557 | 0.0218 | Yes |
| 2 | ADI1 | NA | 1253 | 0.507 | 0.0450 | Yes |
| 3 | CTH | NA | 1775 | 0.469 | 0.0654 | Yes |
| 4 | MTHFD2L | NA | 1836 | 0.466 | 0.0941 | Yes |
| 5 | NAT8L | NA | 1963 | 0.458 | 0.1211 | Yes |
| 6 | BCAT2 | NA | 2691 | 0.422 | 0.1347 | Yes |
| 7 | SRR | NA | 3405 | 0.394 | 0.1468 | Yes |
| 8 | PSPH | NA | 3679 | 0.384 | 0.1664 | Yes |
| 9 | AASS | NA | 4720 | 0.353 | 0.1699 | Yes |
| 10 | OAT | NA | 4925 | 0.347 | 0.1883 | Yes |
| 11 | SERINC3 | NA | 5046 | 0.344 | 0.2081 | Yes |
| 12 | CBS | NA | 5399 | 0.334 | 0.2230 | Yes |
| 13 | AASDHPPT | NA | 5663 | 0.328 | 0.2392 | Yes |
| 14 | MTR | NA | 5816 | 0.325 | 0.2571 | Yes |
| 15 | APIP | NA | 6386 | 0.312 | 0.2666 | Yes |
| 16 | DHFRP1 | NA | 6709 | 0.305 | 0.2802 | Yes |
| 17 | ALDH18A1 | NA | 6807 | 0.303 | 0.2978 | Yes |
| 18 | SHMT1 | NA | 7155 | 0.296 | 0.3103 | Yes |
| 19 | MTAP | NA | 7426 | 0.290 | 0.3239 | Yes |
| 20 | DHFR | NA | 7628 | 0.286 | 0.3385 | Yes |
| 21 | PLOD3 | NA | 7684 | 0.285 | 0.3558 | Yes |
| 22 | RIMKLA | NA | 8201 | 0.275 | 0.3639 | Yes |
| 23 | GLUD1 | NA | 8290 | 0.274 | 0.3797 | Yes |
| 24 | ACCS | NA | 8623 | 0.268 | 0.3908 | Yes |
| 25 | GLS | NA | 8664 | 0.267 | 0.4071 | Yes |
| 26 | GPT2 | NA | 8854 | 0.264 | 0.4205 | Yes |
| 27 | DHFR2 | NA | 8964 | 0.262 | 0.4353 | Yes |
| 28 | ASL | NA | 9394 | 0.254 | 0.4436 | Yes |
| 29 | PSPHP1 | NA | 9846 | 0.247 | 0.4512 | Yes |
| 30 | NOXRED1 | NA | 10479 | 0.237 | 0.4547 | Yes |
| 31 | SERINC5 | NA | 10552 | 0.236 | 0.4684 | Yes |
| 32 | KYAT1 | NA | 10869 | 0.231 | 0.4774 | Yes |
| 33 | GPT | NA | 11569 | 0.221 | 0.4787 | Yes |
| 34 | MTRR | NA | 11668 | 0.219 | 0.4909 | Yes |
| 35 | ASS1 | NA | 11856 | 0.216 | 0.5012 | Yes |
| 36 | PYCR1 | NA | 11910 | 0.216 | 0.5141 | Yes |
| 37 | BHMT2 | NA | 12202 | 0.212 | 0.5223 | Yes |
| 38 | ILVBL | NA | 12916 | 0.202 | 0.5221 | Yes |
| 39 | ENOPH1 | NA | 13314 | 0.196 | 0.5273 | Yes |
| 40 | SLC25A12 | NA | 13434 | 0.194 | 0.5376 | Yes |
| 41 | PLOD2 | NA | 13626 | 0.192 | 0.5463 | Yes |
| 42 | GGT1 | NA | 14045 | 0.186 | 0.5505 | Yes |
| 43 | BHMT | NA | 14055 | 0.186 | 0.5623 | Yes |
| 44 | ASPG | NA | 16523 | 0.155 | 0.5269 | No |
| 45 | PAH | NA | 16594 | 0.154 | 0.5355 | No |
| 46 | PYCR2 | NA | 16931 | 0.150 | 0.5389 | No |
| 47 | BCAT1 | NA | 17329 | 0.145 | 0.5409 | No |
| 48 | GLUL | NA | 18517 | 0.130 | 0.5274 | No |
| 49 | CLN3 | NA | 18819 | 0.126 | 0.5300 | No |
| 50 | FOLH1B | NA | 18826 | 0.126 | 0.5379 | No |
| 51 | PHGDH | NA | 19837 | 0.113 | 0.5266 | No |
| 52 | GOT2 | NA | 20824 | 0.102 | 0.5150 | No |
| 53 | PARK7 | NA | 20966 | 0.100 | 0.5188 | No |
| 54 | MRI1 | NA | 21090 | 0.098 | 0.5228 | No |
| 55 | AGXT2 | NA | 21376 | 0.095 | 0.5237 | No |
| 56 | GLS2 | NA | 22103 | 0.087 | 0.5159 | No |
| 57 | FOLH1 | NA | 23584 | 0.071 | 0.4933 | No |
| 58 | SHMT2 | NA | 25168 | 0.056 | 0.4678 | No |
| 59 | ASNS | NA | 25196 | 0.055 | 0.4709 | No |
| 60 | GAD1 | NA | 27427 | 0.036 | 0.4323 | No |
| 61 | GOT1 | NA | 27470 | 0.036 | 0.4338 | No |
| 62 | MTHFR | NA | 28420 | 0.030 | 0.4183 | No |
| 63 | DPYD | NA | 29468 | 0.023 | 0.4006 | No |
| 64 | GLUD2 | NA | 29800 | 0.021 | 0.3959 | No |
| 65 | NAALAD2 | NA | 32582 | 0.008 | 0.3455 | No |
| 66 | ASPA | NA | 33724 | 0.003 | 0.3247 | No |
| 67 | OTC | NA | 39269 | -0.002 | 0.2232 | No |
| 68 | NAGS | NA | 39434 | -0.004 | 0.2204 | No |
| 69 | RIMKLB | NA | 43150 | -0.050 | 0.1555 | No |
| 70 | CBSL | NA | 43828 | -0.060 | 0.1469 | No |
| 71 | ASNSD1 | NA | 44545 | -0.070 | 0.1382 | No |
| 72 | PSAT1 | NA | 45914 | -0.090 | 0.1189 | No |
| 73 | GOT1L1 | NA | 47773 | -0.121 | 0.0925 | No |
| 74 | UPB1 | NA | 48522 | -0.135 | 0.0875 | No |
| 75 | AGXT | NA | 48565 | -0.136 | 0.0954 | No |
| 76 | GADL1 | NA | 52773 | -0.237 | 0.0333 | No |