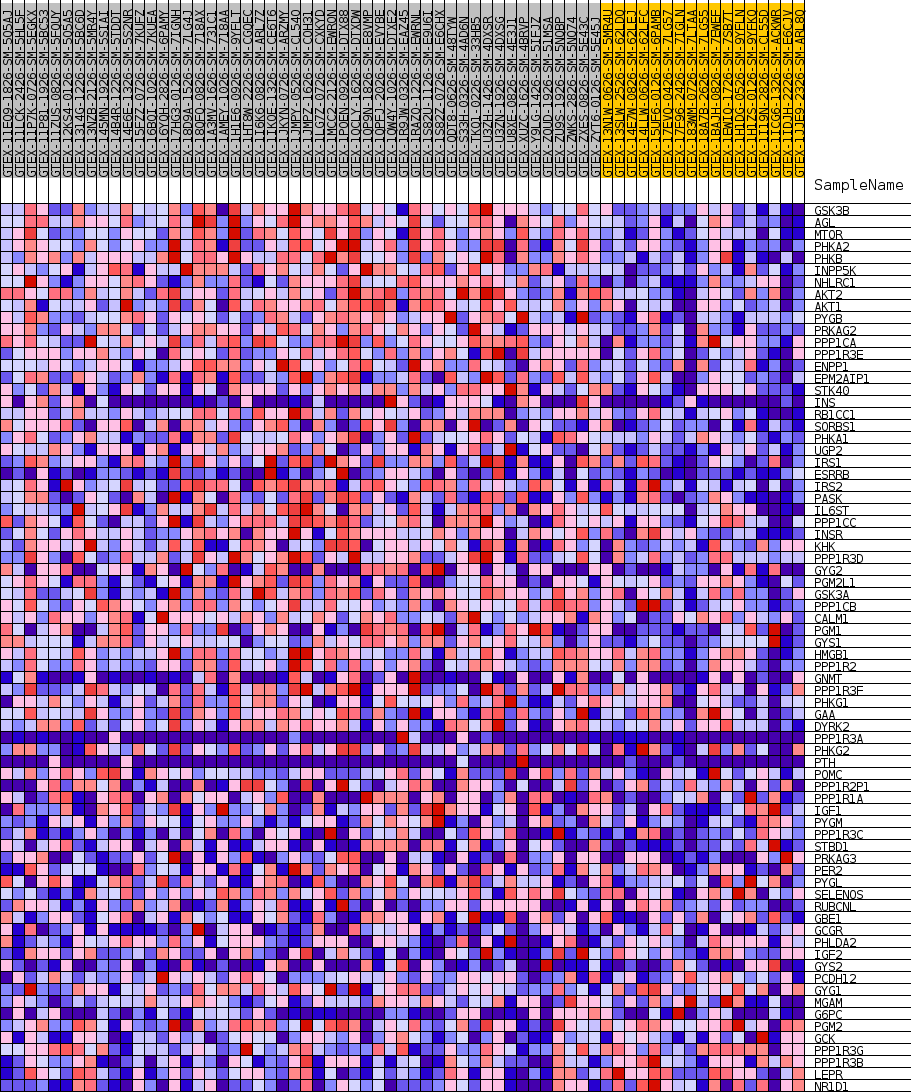
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
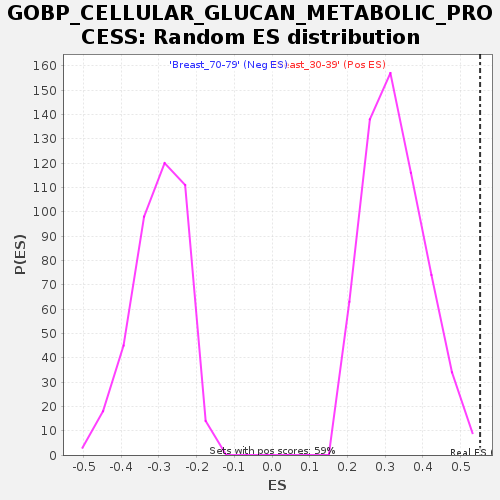
| GeneSet | GOBP_CELLULAR_GLUCAN_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.5519594 |
| Normalized Enrichment Score (NES) | 1.6875365 |
| Nominal p-value | 0.0016920473 |
| FDR q-value | 0.30114785 |
| FWER p-Value | 0.691 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | GSK3B | NA | 325 | 0.632 | 0.0287 | Yes |
| 2 | AGL | NA | 348 | 0.625 | 0.0626 | Yes |
| 3 | MTOR | NA | 355 | 0.623 | 0.0968 | Yes |
| 4 | PHKA2 | NA | 522 | 0.590 | 0.1261 | Yes |
| 5 | PHKB | NA | 549 | 0.585 | 0.1578 | Yes |
| 6 | INPP5K | NA | 1017 | 0.528 | 0.1782 | Yes |
| 7 | NHLRC1 | NA | 1139 | 0.517 | 0.2044 | Yes |
| 8 | AKT2 | NA | 1358 | 0.498 | 0.2278 | Yes |
| 9 | AKT1 | NA | 1390 | 0.496 | 0.2545 | Yes |
| 10 | PYGB | NA | 1603 | 0.482 | 0.2771 | Yes |
| 11 | PRKAG2 | NA | 1741 | 0.473 | 0.3005 | Yes |
| 12 | PPP1CA | NA | 3859 | 0.378 | 0.2825 | Yes |
| 13 | PPP1R3E | NA | 4297 | 0.365 | 0.2945 | Yes |
| 14 | ENPP1 | NA | 4763 | 0.352 | 0.3053 | Yes |
| 15 | EPM2AIP1 | NA | 5024 | 0.345 | 0.3195 | Yes |
| 16 | STK40 | NA | 5186 | 0.340 | 0.3352 | Yes |
| 17 | INS | NA | 5395 | 0.335 | 0.3498 | Yes |
| 18 | RB1CC1 | NA | 5479 | 0.333 | 0.3665 | Yes |
| 19 | SORBS1 | NA | 5768 | 0.326 | 0.3792 | Yes |
| 20 | PHKA1 | NA | 6009 | 0.320 | 0.3923 | Yes |
| 21 | UGP2 | NA | 6459 | 0.310 | 0.4011 | Yes |
| 22 | IRS1 | NA | 6700 | 0.305 | 0.4135 | Yes |
| 23 | ESRRB | NA | 6959 | 0.300 | 0.4252 | Yes |
| 24 | IRS2 | NA | 7052 | 0.298 | 0.4399 | Yes |
| 25 | PASK | NA | 7442 | 0.290 | 0.4487 | Yes |
| 26 | IL6ST | NA | 7499 | 0.289 | 0.4635 | Yes |
| 27 | PPP1CC | NA | 8264 | 0.274 | 0.4646 | Yes |
| 28 | INSR | NA | 9620 | 0.250 | 0.4535 | Yes |
| 29 | KHK | NA | 9764 | 0.248 | 0.4645 | Yes |
| 30 | PPP1R3D | NA | 10134 | 0.242 | 0.4710 | Yes |
| 31 | GYG2 | NA | 10248 | 0.240 | 0.4822 | Yes |
| 32 | PGM2L1 | NA | 10942 | 0.230 | 0.4821 | Yes |
| 33 | GSK3A | NA | 11015 | 0.229 | 0.4933 | Yes |
| 34 | PPP1CB | NA | 11041 | 0.228 | 0.5054 | Yes |
| 35 | CALM1 | NA | 11085 | 0.228 | 0.5171 | Yes |
| 36 | PGM1 | NA | 11277 | 0.225 | 0.5260 | Yes |
| 37 | GYS1 | NA | 11300 | 0.225 | 0.5379 | Yes |
| 38 | HMGB1 | NA | 11903 | 0.216 | 0.5387 | Yes |
| 39 | PPP1R2 | NA | 12464 | 0.208 | 0.5399 | Yes |
| 40 | GNMT | NA | 12793 | 0.204 | 0.5451 | Yes |
| 41 | PPP1R3F | NA | 13020 | 0.201 | 0.5520 | Yes |
| 42 | PHKG1 | NA | 13875 | 0.188 | 0.5466 | No |
| 43 | GAA | NA | 14244 | 0.183 | 0.5500 | No |
| 44 | DYRK2 | NA | 15561 | 0.167 | 0.5350 | No |
| 45 | PPP1R3A | NA | 16023 | 0.161 | 0.5354 | No |
| 46 | PHKG2 | NA | 17441 | 0.144 | 0.5173 | No |
| 47 | PTH | NA | 17567 | 0.142 | 0.5228 | No |
| 48 | POMC | NA | 18890 | 0.125 | 0.5054 | No |
| 49 | PPP1R2P1 | NA | 20917 | 0.101 | 0.4738 | No |
| 50 | PPP1R1A | NA | 21236 | 0.097 | 0.4733 | No |
| 51 | IGF1 | NA | 24322 | 0.064 | 0.4202 | No |
| 52 | PYGM | NA | 25568 | 0.052 | 0.4002 | No |
| 53 | PPP1R3C | NA | 25837 | 0.049 | 0.3980 | No |
| 54 | STBD1 | NA | 26285 | 0.045 | 0.3923 | No |
| 55 | PRKAG3 | NA | 26374 | 0.044 | 0.3931 | No |
| 56 | PER2 | NA | 26679 | 0.042 | 0.3898 | No |
| 57 | PYGL | NA | 26759 | 0.041 | 0.3906 | No |
| 58 | SELENOS | NA | 29430 | 0.024 | 0.3430 | No |
| 59 | RUBCNL | NA | 29744 | 0.022 | 0.3384 | No |
| 60 | GBE1 | NA | 32851 | 0.007 | 0.2819 | No |
| 61 | GCGR | NA | 34011 | 0.000 | 0.2606 | No |
| 62 | PHLDA2 | NA | 39302 | -0.002 | 0.1637 | No |
| 63 | IGF2 | NA | 40933 | -0.023 | 0.1351 | No |
| 64 | GYS2 | NA | 41872 | -0.034 | 0.1197 | No |
| 65 | PCDH12 | NA | 42589 | -0.043 | 0.1090 | No |
| 66 | GYG1 | NA | 46324 | -0.096 | 0.0458 | No |
| 67 | MGAM | NA | 49361 | -0.152 | -0.0015 | No |
| 68 | G6PC | NA | 49768 | -0.161 | -0.0001 | No |
| 69 | PGM2 | NA | 50318 | -0.173 | -0.0007 | No |
| 70 | GCK | NA | 51070 | -0.191 | -0.0040 | No |
| 71 | PPP1R3G | NA | 51169 | -0.193 | 0.0048 | No |
| 72 | PPP1R3B | NA | 53782 | -0.300 | -0.0266 | No |
| 73 | LEPR | NA | 54108 | -0.343 | -0.0137 | No |
| 74 | NR1D1 | NA | 54350 | -0.410 | 0.0044 | No |