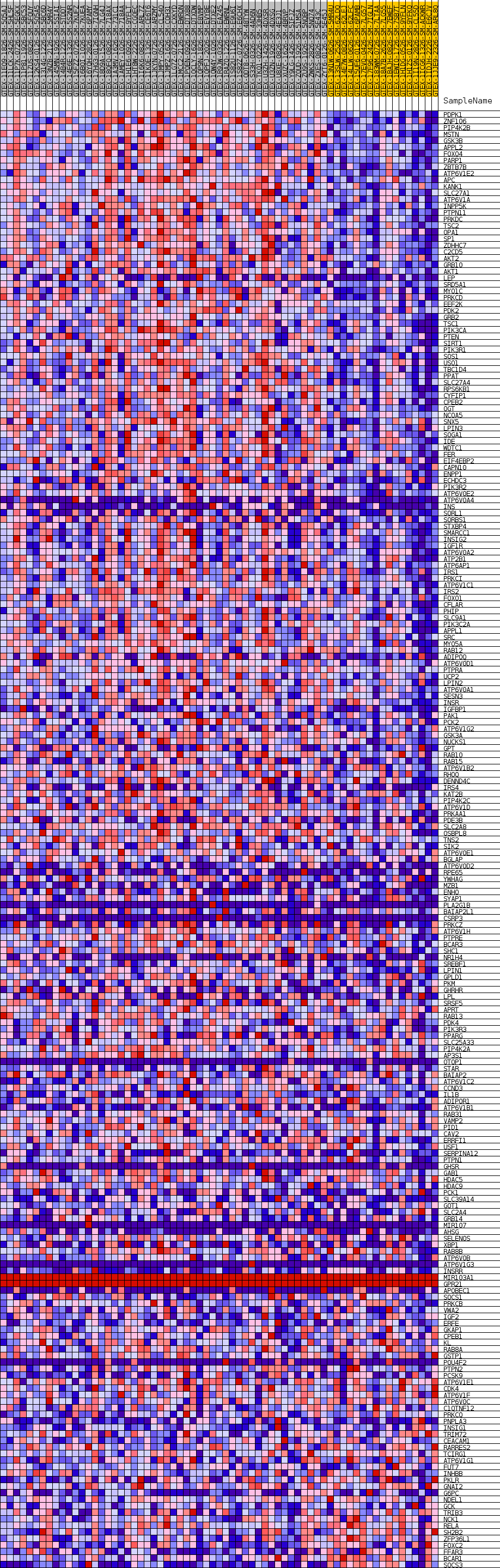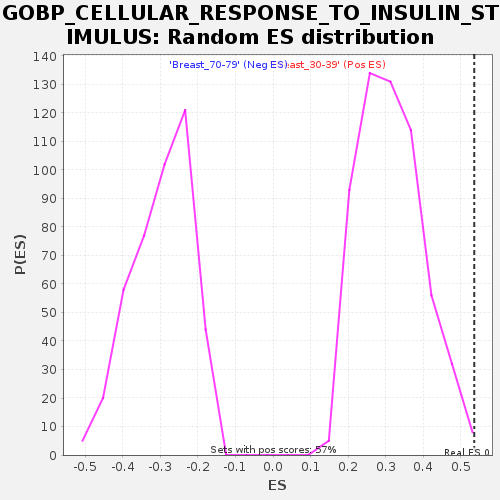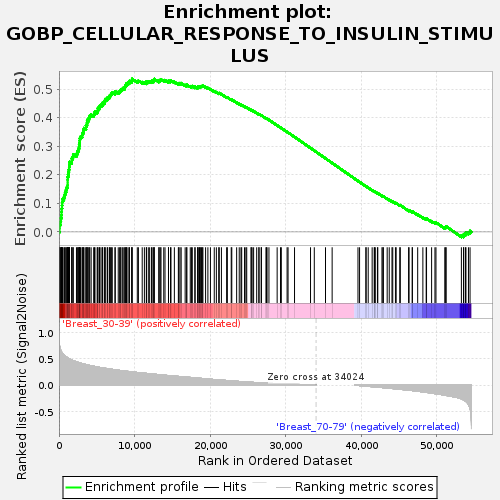
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_CELLULAR_RESPONSE_TO_INSULIN_STIMULUS |
| Enrichment Score (ES) | 0.53533465 |
| Normalized Enrichment Score (NES) | 1.6983054 |
| Nominal p-value | 0.0017452007 |
| FDR q-value | 0.31498954 |
| FWER p-Value | 0.653 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | PDPK1 | NA | 76 | 0.745 | 0.0126 | Yes |
| 2 | ZNF106 | NA | 78 | 0.743 | 0.0266 | Yes |
| 3 | PIP4K2B | NA | 221 | 0.663 | 0.0365 | Yes |
| 4 | MSTN | NA | 238 | 0.657 | 0.0485 | Yes |
| 5 | GSK3B | NA | 325 | 0.632 | 0.0588 | Yes |
| 6 | APPL2 | NA | 333 | 0.629 | 0.0705 | Yes |
| 7 | FOXO4 | NA | 359 | 0.623 | 0.0818 | Yes |
| 8 | PARP1 | NA | 404 | 0.613 | 0.0925 | Yes |
| 9 | ZBTB7B | NA | 421 | 0.610 | 0.1037 | Yes |
| 10 | ATP6V1E2 | NA | 454 | 0.603 | 0.1145 | Yes |
| 11 | APC | NA | 645 | 0.570 | 0.1217 | Yes |
| 12 | KANK1 | NA | 740 | 0.558 | 0.1305 | Yes |
| 13 | SLC27A1 | NA | 819 | 0.549 | 0.1393 | Yes |
| 14 | ATP6V1A | NA | 932 | 0.536 | 0.1474 | Yes |
| 15 | INPP5K | NA | 1017 | 0.528 | 0.1558 | Yes |
| 16 | PTPN11 | NA | 1141 | 0.517 | 0.1632 | Yes |
| 17 | PRKDC | NA | 1146 | 0.516 | 0.1729 | Yes |
| 18 | TSC2 | NA | 1147 | 0.516 | 0.1826 | Yes |
| 19 | OPA1 | NA | 1153 | 0.516 | 0.1922 | Yes |
| 20 | SP1 | NA | 1208 | 0.512 | 0.2009 | Yes |
| 21 | ZDHHC7 | NA | 1258 | 0.507 | 0.2095 | Yes |
| 22 | C2CD5 | NA | 1305 | 0.503 | 0.2181 | Yes |
| 23 | AKT2 | NA | 1358 | 0.498 | 0.2265 | Yes |
| 24 | GRB10 | NA | 1373 | 0.497 | 0.2356 | Yes |
| 25 | AKT1 | NA | 1390 | 0.496 | 0.2447 | Yes |
| 26 | LEP | NA | 1684 | 0.477 | 0.2483 | Yes |
| 27 | SRD5A1 | NA | 1687 | 0.476 | 0.2572 | Yes |
| 28 | MYO1C | NA | 1832 | 0.466 | 0.2633 | Yes |
| 29 | PRKCD | NA | 1888 | 0.463 | 0.2710 | Yes |
| 30 | EEF2K | NA | 2311 | 0.440 | 0.2715 | Yes |
| 31 | PDK2 | NA | 2412 | 0.435 | 0.2779 | Yes |
| 32 | GRB2 | NA | 2497 | 0.431 | 0.2845 | Yes |
| 33 | TSC1 | NA | 2614 | 0.426 | 0.2903 | Yes |
| 34 | PIK3CA | NA | 2652 | 0.424 | 0.2976 | Yes |
| 35 | PTEN | NA | 2673 | 0.423 | 0.3052 | Yes |
| 36 | SIRT1 | NA | 2723 | 0.421 | 0.3123 | Yes |
| 37 | PIK3R1 | NA | 2739 | 0.420 | 0.3199 | Yes |
| 38 | SOS1 | NA | 2754 | 0.420 | 0.3275 | Yes |
| 39 | USO1 | NA | 2853 | 0.415 | 0.3335 | Yes |
| 40 | TBC1D4 | NA | 3061 | 0.407 | 0.3374 | Yes |
| 41 | PPAT | NA | 3087 | 0.406 | 0.3446 | Yes |
| 42 | SLC27A4 | NA | 3199 | 0.402 | 0.3501 | Yes |
| 43 | RPS6KB1 | NA | 3228 | 0.401 | 0.3571 | Yes |
| 44 | CYFIP1 | NA | 3310 | 0.398 | 0.3631 | Yes |
| 45 | CPEB2 | NA | 3565 | 0.389 | 0.3658 | Yes |
| 46 | OGT | NA | 3581 | 0.388 | 0.3728 | Yes |
| 47 | NCOA5 | NA | 3699 | 0.384 | 0.3779 | Yes |
| 48 | SNX5 | NA | 3720 | 0.383 | 0.3847 | Yes |
| 49 | LPIN3 | NA | 3734 | 0.382 | 0.3917 | Yes |
| 50 | SOGA1 | NA | 3947 | 0.375 | 0.3948 | Yes |
| 51 | IDE | NA | 3980 | 0.375 | 0.4013 | Yes |
| 52 | WDTC1 | NA | 4066 | 0.372 | 0.4067 | Yes |
| 53 | FER | NA | 4254 | 0.367 | 0.4102 | Yes |
| 54 | EIF4EBP2 | NA | 4590 | 0.357 | 0.4107 | Yes |
| 55 | CAPN10 | NA | 4702 | 0.353 | 0.4154 | Yes |
| 56 | ENPP1 | NA | 4763 | 0.352 | 0.4209 | Yes |
| 57 | ECHDC3 | NA | 5052 | 0.344 | 0.4221 | Yes |
| 58 | PIK3R2 | NA | 5143 | 0.342 | 0.4268 | Yes |
| 59 | ATP6V0E2 | NA | 5146 | 0.342 | 0.4332 | Yes |
| 60 | ATP6V0A4 | NA | 5361 | 0.335 | 0.4356 | Yes |
| 61 | INS | NA | 5395 | 0.335 | 0.4413 | Yes |
| 62 | SORL1 | NA | 5570 | 0.331 | 0.4443 | Yes |
| 63 | SORBS1 | NA | 5768 | 0.326 | 0.4468 | Yes |
| 64 | STXBP4 | NA | 5781 | 0.326 | 0.4527 | Yes |
| 65 | SMARCC1 | NA | 6037 | 0.320 | 0.4541 | Yes |
| 66 | INSIG2 | NA | 6072 | 0.319 | 0.4594 | Yes |
| 67 | IGF1R | NA | 6119 | 0.318 | 0.4646 | Yes |
| 68 | ATP6V0A2 | NA | 6379 | 0.312 | 0.4657 | Yes |
| 69 | ATP2B1 | NA | 6429 | 0.311 | 0.4706 | Yes |
| 70 | ATP6AP1 | NA | 6657 | 0.306 | 0.4722 | Yes |
| 71 | IRS1 | NA | 6700 | 0.305 | 0.4772 | Yes |
| 72 | PRKCI | NA | 6833 | 0.302 | 0.4804 | Yes |
| 73 | ATP6V1C1 | NA | 6933 | 0.300 | 0.4843 | Yes |
| 74 | IRS2 | NA | 7052 | 0.298 | 0.4877 | Yes |
| 75 | FOXO1 | NA | 7408 | 0.291 | 0.4866 | Yes |
| 76 | CFLAR | NA | 7446 | 0.290 | 0.4914 | Yes |
| 77 | PHIP | NA | 7842 | 0.282 | 0.4895 | Yes |
| 78 | SLC9A1 | NA | 8027 | 0.278 | 0.4913 | Yes |
| 79 | PIK3C2A | NA | 8112 | 0.277 | 0.4950 | Yes |
| 80 | APPL1 | NA | 8192 | 0.275 | 0.4987 | Yes |
| 81 | SRC | NA | 8379 | 0.272 | 0.5004 | Yes |
| 82 | MYO5A | NA | 8496 | 0.270 | 0.5034 | Yes |
| 83 | RAB12 | NA | 8709 | 0.267 | 0.5045 | Yes |
| 84 | ADIPOQ | NA | 8737 | 0.266 | 0.5090 | Yes |
| 85 | ATP6V0D1 | NA | 8790 | 0.265 | 0.5130 | Yes |
| 86 | PTPRA | NA | 8928 | 0.263 | 0.5154 | Yes |
| 87 | UCP2 | NA | 8945 | 0.262 | 0.5201 | Yes |
| 88 | LPIN2 | NA | 9132 | 0.259 | 0.5215 | Yes |
| 89 | ATP6V0A1 | NA | 9309 | 0.256 | 0.5231 | Yes |
| 90 | SESN3 | NA | 9310 | 0.256 | 0.5279 | Yes |
| 91 | INSR | NA | 9620 | 0.250 | 0.5270 | Yes |
| 92 | IGFBP1 | NA | 9667 | 0.250 | 0.5308 | Yes |
| 93 | PAK1 | NA | 9677 | 0.250 | 0.5353 | Yes |
| 94 | PCK2 | NA | 10364 | 0.239 | 0.5272 | No |
| 95 | ATP6V1G2 | NA | 10512 | 0.236 | 0.5289 | No |
| 96 | GSK3A | NA | 11015 | 0.229 | 0.5240 | No |
| 97 | NUCKS1 | NA | 11303 | 0.224 | 0.5230 | No |
| 98 | GPT | NA | 11569 | 0.221 | 0.5222 | No |
| 99 | RAB10 | NA | 11572 | 0.221 | 0.5264 | No |
| 100 | RAB15 | NA | 11822 | 0.217 | 0.5259 | No |
| 101 | ATP6V1B2 | NA | 12000 | 0.215 | 0.5266 | No |
| 102 | RHOQ | NA | 12252 | 0.211 | 0.5260 | No |
| 103 | DENND4C | NA | 12323 | 0.210 | 0.5287 | No |
| 104 | IRS4 | NA | 12543 | 0.207 | 0.5286 | No |
| 105 | KAT2B | NA | 12557 | 0.207 | 0.5322 | No |
| 106 | PIP4K2C | NA | 12621 | 0.206 | 0.5349 | No |
| 107 | ATP6V1D | NA | 13206 | 0.198 | 0.5279 | No |
| 108 | PRKAA1 | NA | 13261 | 0.197 | 0.5306 | No |
| 109 | PDE3B | NA | 13402 | 0.195 | 0.5317 | No |
| 110 | SLC2A8 | NA | 13504 | 0.193 | 0.5335 | No |
| 111 | OSBPL8 | NA | 13867 | 0.188 | 0.5304 | No |
| 112 | TNS2 | NA | 14033 | 0.186 | 0.5309 | No |
| 113 | SIK2 | NA | 14492 | 0.180 | 0.5258 | No |
| 114 | ATP6V0E1 | NA | 14496 | 0.180 | 0.5292 | No |
| 115 | BGLAP | NA | 14760 | 0.177 | 0.5276 | No |
| 116 | ATP6V0D2 | NA | 14824 | 0.176 | 0.5298 | No |
| 117 | RPE65 | NA | 15271 | 0.170 | 0.5248 | No |
| 118 | YWHAG | NA | 15800 | 0.164 | 0.5182 | No |
| 119 | MZB1 | NA | 15862 | 0.163 | 0.5201 | No |
| 120 | ENHO | NA | 16019 | 0.161 | 0.5203 | No |
| 121 | SYAP1 | NA | 16173 | 0.159 | 0.5205 | No |
| 122 | PLA2G1B | NA | 16700 | 0.153 | 0.5137 | No |
| 123 | BAIAP2L1 | NA | 16910 | 0.150 | 0.5127 | No |
| 124 | CSRP3 | NA | 16911 | 0.150 | 0.5155 | No |
| 125 | PRKCZ | NA | 17376 | 0.145 | 0.5097 | No |
| 126 | ATP6V1H | NA | 17534 | 0.143 | 0.5095 | No |
| 127 | PTPRE | NA | 17645 | 0.141 | 0.5101 | No |
| 128 | BCAR3 | NA | 17983 | 0.137 | 0.5065 | No |
| 129 | SHC1 | NA | 18018 | 0.136 | 0.5084 | No |
| 130 | NR1H4 | NA | 18353 | 0.132 | 0.5048 | No |
| 131 | SREBF1 | NA | 18357 | 0.132 | 0.5072 | No |
| 132 | LPIN1 | NA | 18498 | 0.130 | 0.5071 | No |
| 133 | GPLD1 | NA | 18598 | 0.129 | 0.5077 | No |
| 134 | PKM | NA | 18662 | 0.128 | 0.5089 | No |
| 135 | GHRHR | NA | 18759 | 0.127 | 0.5095 | No |
| 136 | LPL | NA | 18871 | 0.125 | 0.5099 | No |
| 137 | SRSF5 | NA | 18975 | 0.124 | 0.5103 | No |
| 138 | APRT | NA | 19052 | 0.123 | 0.5112 | No |
| 139 | RAB13 | NA | 19405 | 0.119 | 0.5070 | No |
| 140 | PDK4 | NA | 19710 | 0.115 | 0.5036 | No |
| 141 | PIK3R3 | NA | 20047 | 0.111 | 0.4995 | No |
| 142 | PPARG | NA | 20553 | 0.105 | 0.4921 | No |
| 143 | SLC25A33 | NA | 20831 | 0.102 | 0.4890 | No |
| 144 | PIP4K2A | NA | 21146 | 0.098 | 0.4850 | No |
| 145 | AP3S1 | NA | 21213 | 0.097 | 0.4856 | No |
| 146 | OTOP1 | NA | 21500 | 0.094 | 0.4821 | No |
| 147 | STAR | NA | 22191 | 0.086 | 0.4711 | No |
| 148 | BAIAP2 | NA | 22286 | 0.085 | 0.4709 | No |
| 149 | ATP6V1C2 | NA | 22798 | 0.079 | 0.4630 | No |
| 150 | CCND3 | NA | 22874 | 0.079 | 0.4631 | No |
| 151 | IL1B | NA | 23517 | 0.072 | 0.4527 | No |
| 152 | ADIPOR1 | NA | 23859 | 0.068 | 0.4477 | No |
| 153 | ATP6V1B1 | NA | 24089 | 0.066 | 0.4447 | No |
| 154 | RAB31 | NA | 24184 | 0.065 | 0.4442 | No |
| 155 | VAMP2 | NA | 24560 | 0.061 | 0.4385 | No |
| 156 | PID1 | NA | 24739 | 0.060 | 0.4363 | No |
| 157 | CAV2 | NA | 24851 | 0.059 | 0.4354 | No |
| 158 | ERRFI1 | NA | 25430 | 0.053 | 0.4258 | No |
| 159 | USF1 | NA | 25448 | 0.053 | 0.4264 | No |
| 160 | SERPINA12 | NA | 25564 | 0.052 | 0.4253 | No |
| 161 | PTPN1 | NA | 25762 | 0.050 | 0.4226 | No |
| 162 | GHSR | NA | 26170 | 0.046 | 0.4160 | No |
| 163 | GAB1 | NA | 26457 | 0.044 | 0.4116 | No |
| 164 | HDAC5 | NA | 26481 | 0.043 | 0.4119 | No |
| 165 | HDAC9 | NA | 26764 | 0.041 | 0.4075 | No |
| 166 | PCK1 | NA | 26802 | 0.041 | 0.4076 | No |
| 167 | SLC39A14 | NA | 27398 | 0.037 | 0.3974 | No |
| 168 | GOT1 | NA | 27470 | 0.036 | 0.3967 | No |
| 169 | SLC2A4 | NA | 27543 | 0.036 | 0.3961 | No |
| 170 | GRB14 | NA | 27793 | 0.034 | 0.3922 | No |
| 171 | MIR107 | NA | 28878 | 0.027 | 0.3727 | No |
| 172 | AHSG | NA | 29322 | 0.024 | 0.3650 | No |
| 173 | SELENOS | NA | 29430 | 0.024 | 0.3635 | No |
| 174 | XBP1 | NA | 30195 | 0.019 | 0.3498 | No |
| 175 | RAB8B | NA | 30323 | 0.019 | 0.3478 | No |
| 176 | ATP6V0B | NA | 31179 | 0.014 | 0.3324 | No |
| 177 | ATP6V1G3 | NA | 33297 | 0.006 | 0.2935 | No |
| 178 | INSRR | NA | 33795 | 0.003 | 0.2844 | No |
| 179 | MIR103A1 | NA | 35279 | 0.000 | 0.2572 | No |
| 180 | GPR21 | NA | 36158 | 0.000 | 0.2410 | No |
| 181 | APOBEC1 | NA | 39579 | -0.006 | 0.1782 | No |
| 182 | SOCS1 | NA | 39789 | -0.009 | 0.1746 | No |
| 183 | PRKCB | NA | 40625 | -0.019 | 0.1596 | No |
| 184 | VWA2 | NA | 40654 | -0.020 | 0.1594 | No |
| 185 | IGF2 | NA | 40933 | -0.023 | 0.1547 | No |
| 186 | ERFE | NA | 41455 | -0.029 | 0.1457 | No |
| 187 | GKAP1 | NA | 41733 | -0.032 | 0.1412 | No |
| 188 | CPEB1 | NA | 41759 | -0.033 | 0.1414 | No |
| 189 | KL | NA | 41871 | -0.034 | 0.1400 | No |
| 190 | RAB8A | NA | 42154 | -0.037 | 0.1355 | No |
| 191 | GSTP1 | NA | 42202 | -0.038 | 0.1353 | No |
| 192 | POU4F2 | NA | 42754 | -0.045 | 0.1260 | No |
| 193 | PTPN2 | NA | 42872 | -0.047 | 0.1248 | No |
| 194 | PCSK9 | NA | 42943 | -0.048 | 0.1244 | No |
| 195 | ATP6V1E1 | NA | 43444 | -0.054 | 0.1162 | No |
| 196 | CDK4 | NA | 43717 | -0.058 | 0.1123 | No |
| 197 | ATP6V1F | NA | 44080 | -0.063 | 0.1068 | No |
| 198 | ATP6V0C | NA | 44182 | -0.065 | 0.1062 | No |
| 199 | C1QTNF12 | NA | 44539 | -0.070 | 0.1009 | No |
| 200 | PRKCQ | NA | 44633 | -0.071 | 0.1006 | No |
| 201 | PNPLA3 | NA | 45113 | -0.078 | 0.0932 | No |
| 202 | INSIG1 | NA | 45178 | -0.079 | 0.0935 | No |
| 203 | TRIM72 | NA | 46278 | -0.096 | 0.0751 | No |
| 204 | CEACAM1 | NA | 46340 | -0.097 | 0.0758 | No |
| 205 | RARRES2 | NA | 46763 | -0.103 | 0.0700 | No |
| 206 | TCIRG1 | NA | 46776 | -0.104 | 0.0717 | No |
| 207 | ATP6V1G1 | NA | 47483 | -0.116 | 0.0609 | No |
| 208 | FUT7 | NA | 48169 | -0.129 | 0.0508 | No |
| 209 | INHBB | NA | 48608 | -0.137 | 0.0453 | No |
| 210 | PKLR | NA | 48654 | -0.138 | 0.0470 | No |
| 211 | GNAI2 | NA | 49326 | -0.151 | 0.0376 | No |
| 212 | G6PC | NA | 49768 | -0.161 | 0.0325 | No |
| 213 | NDEL1 | NA | 49903 | -0.164 | 0.0331 | No |
| 214 | GCK | NA | 51070 | -0.191 | 0.0152 | No |
| 215 | TRIB3 | NA | 51182 | -0.194 | 0.0168 | No |
| 216 | NCK1 | NA | 51252 | -0.196 | 0.0193 | No |
| 217 | RELA | NA | 53267 | -0.259 | -0.0129 | No |
| 218 | SH2B2 | NA | 53549 | -0.278 | -0.0129 | No |
| 219 | ZFP36L1 | NA | 53580 | -0.281 | -0.0081 | No |
| 220 | FOXC2 | NA | 53809 | -0.302 | -0.0066 | No |
| 221 | FFAR3 | NA | 53872 | -0.308 | -0.0020 | No |
| 222 | BCAR1 | NA | 54204 | -0.364 | -0.0012 | No |
| 223 | SOCS3 | NA | 54415 | -0.440 | 0.0032 | No |