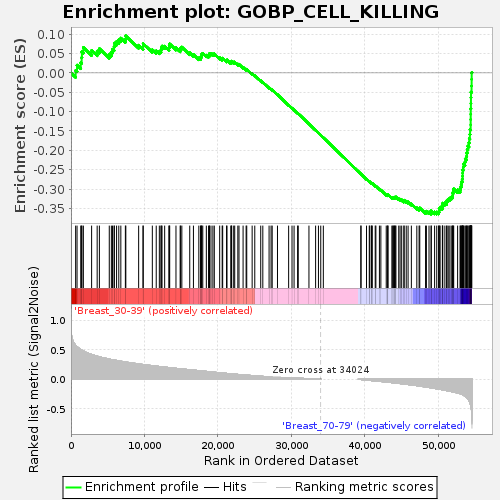
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_70-79 |
| GeneSet | GOBP_CELL_KILLING |
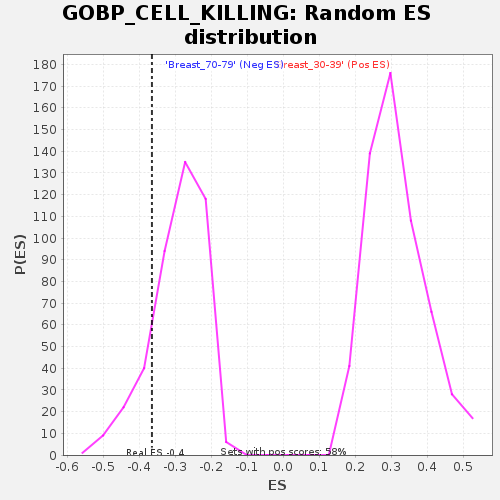
| Enrichment Score (ES) | -0.3644929 |
| Normalized Enrichment Score (NES) | -1.2424443 |
| Nominal p-value | 0.16235293 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.998 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | STAT5B | NA | 632 | 0.572 | 0.0063 | No |
| 2 | TUSC2 | NA | 822 | 0.548 | 0.0200 | No |
| 3 | PPP3CB | NA | 1327 | 0.501 | 0.0264 | No |
| 4 | CD160 | NA | 1447 | 0.492 | 0.0396 | No |
| 5 | AP1G1 | NA | 1483 | 0.489 | 0.0543 | No |
| 6 | LEP | NA | 1684 | 0.477 | 0.0656 | No |
| 7 | DNASE1 | NA | 2807 | 0.417 | 0.0580 | No |
| 8 | CRK | NA | 3562 | 0.389 | 0.0563 | No |
| 9 | VAMP7 | NA | 3864 | 0.378 | 0.0626 | No |
| 10 | BCL2L11 | NA | 5211 | 0.339 | 0.0485 | No |
| 11 | SLC22A13 | NA | 5494 | 0.332 | 0.0538 | No |
| 12 | CLEC7A | NA | 5655 | 0.329 | 0.0611 | No |
| 13 | CEBPG | NA | 5871 | 0.323 | 0.0673 | No |
| 14 | STX7 | NA | 5899 | 0.323 | 0.0769 | No |
| 15 | CD226 | NA | 6206 | 0.316 | 0.0812 | No |
| 16 | MR1 | NA | 6506 | 0.309 | 0.0853 | No |
| 17 | CADM1 | NA | 6785 | 0.303 | 0.0897 | No |
| 18 | FADD | NA | 7402 | 0.291 | 0.0875 | No |
| 19 | IL23A | NA | 7465 | 0.290 | 0.0954 | No |
| 20 | LCE3A | NA | 9211 | 0.258 | 0.0714 | No |
| 21 | TUBB4B | NA | 9807 | 0.248 | 0.0683 | No |
| 22 | TUBB | NA | 9829 | 0.247 | 0.0756 | No |
| 23 | MUC7 | NA | 11073 | 0.228 | 0.0599 | No |
| 24 | REG3G | NA | 11604 | 0.220 | 0.0571 | No |
| 25 | LYZ | NA | 12027 | 0.214 | 0.0560 | No |
| 26 | F2RL1 | NA | 12244 | 0.211 | 0.0587 | No |
| 27 | LAMP1 | NA | 12298 | 0.211 | 0.0643 | No |
| 28 | DEFB118 | NA | 12392 | 0.210 | 0.0692 | No |
| 29 | SYK | NA | 12753 | 0.205 | 0.0689 | No |
| 30 | PTPN6 | NA | 13334 | 0.196 | 0.0644 | No |
| 31 | DEFA6 | NA | 13376 | 0.195 | 0.0698 | No |
| 32 | DEFA5 | NA | 13438 | 0.194 | 0.0747 | No |
| 33 | AGER | NA | 14292 | 0.183 | 0.0648 | No |
| 34 | CXCL6 | NA | 14860 | 0.176 | 0.0599 | No |
| 35 | CASP8 | NA | 14923 | 0.175 | 0.0642 | No |
| 36 | IL7R | NA | 15091 | 0.173 | 0.0665 | No |
| 37 | IL12A | NA | 16167 | 0.159 | 0.0518 | No |
| 38 | EMP2 | NA | 16669 | 0.153 | 0.0474 | No |
| 39 | PGLYRP4 | NA | 17404 | 0.144 | 0.0384 | No |
| 40 | PIK3R6 | NA | 17640 | 0.141 | 0.0385 | No |
| 41 | IL12B | NA | 17687 | 0.141 | 0.0421 | No |
| 42 | SERPINB4 | NA | 17772 | 0.140 | 0.0449 | No |
| 43 | CORO1A | NA | 17805 | 0.139 | 0.0487 | No |
| 44 | CST11 | NA | 17941 | 0.137 | 0.0505 | No |
| 45 | LTF | NA | 18426 | 0.131 | 0.0457 | No |
| 46 | SLAMF6 | NA | 18730 | 0.127 | 0.0441 | No |
| 47 | SLAMF7 | NA | 18841 | 0.126 | 0.0460 | No |
| 48 | DCD | NA | 18844 | 0.126 | 0.0499 | No |
| 49 | CD1E | NA | 19058 | 0.123 | 0.0499 | No |
| 50 | CHGA | NA | 19320 | 0.120 | 0.0488 | No |
| 51 | STXBP2 | NA | 19511 | 0.117 | 0.0490 | No |
| 52 | CD1C | NA | 20243 | 0.108 | 0.0390 | No |
| 53 | SEMG1 | NA | 20583 | 0.105 | 0.0360 | No |
| 54 | CFHR1 | NA | 20652 | 0.104 | 0.0380 | No |
| 55 | HRG | NA | 21179 | 0.098 | 0.0314 | No |
| 56 | CD1B | NA | 21237 | 0.097 | 0.0334 | No |
| 57 | P2RX7 | NA | 21752 | 0.091 | 0.0268 | No |
| 58 | IL23R | NA | 21868 | 0.090 | 0.0275 | No |
| 59 | HAVCR2 | NA | 21869 | 0.090 | 0.0303 | No |
| 60 | FCGR2B | NA | 22153 | 0.087 | 0.0278 | No |
| 61 | DEFB4A | NA | 22290 | 0.085 | 0.0279 | No |
| 62 | CFH | NA | 22715 | 0.080 | 0.0227 | No |
| 63 | BAD | NA | 22872 | 0.079 | 0.0223 | No |
| 64 | KLRF2 | NA | 23430 | 0.073 | 0.0143 | No |
| 65 | XCL1 | NA | 23843 | 0.068 | 0.0089 | No |
| 66 | UNC13D | NA | 23928 | 0.068 | 0.0094 | No |
| 67 | AZGP1 | NA | 24663 | 0.060 | -0.0022 | No |
| 68 | CRTAM | NA | 25021 | 0.057 | -0.0069 | No |
| 69 | RIPK3 | NA | 25831 | 0.049 | -0.0203 | No |
| 70 | ITGAM | NA | 26116 | 0.047 | -0.0240 | No |
| 71 | IL18 | NA | 26968 | 0.040 | -0.0384 | No |
| 72 | LILRB1 | NA | 27240 | 0.038 | -0.0422 | No |
| 73 | CTSG | NA | 27392 | 0.037 | -0.0438 | No |
| 74 | KRT6A | NA | 28113 | 0.032 | -0.0561 | No |
| 75 | LYST | NA | 29633 | 0.022 | -0.0833 | No |
| 76 | DEFB103B | NA | 30091 | 0.020 | -0.0911 | No |
| 77 | CD1A | NA | 30372 | 0.018 | -0.0956 | No |
| 78 | HTN1 | NA | 30852 | 0.016 | -0.1039 | No |
| 79 | HTN3 | NA | 30939 | 0.016 | -0.1050 | No |
| 80 | C9 | NA | 32397 | 0.009 | -0.1315 | No |
| 81 | DEFB128 | NA | 33305 | 0.006 | -0.1480 | No |
| 82 | BCL2L1 | NA | 33690 | 0.003 | -0.1550 | No |
| 83 | LCE3B | NA | 34004 | 0.000 | -0.1607 | No |
| 84 | DEFB103A | NA | 34355 | 0.000 | -0.1671 | No |
| 85 | HAMP | NA | 39468 | -0.005 | -0.2609 | No |
| 86 | KLRC2 | NA | 39484 | -0.005 | -0.2611 | No |
| 87 | LGALS9 | NA | 40236 | -0.015 | -0.2744 | No |
| 88 | STAP1 | NA | 40610 | -0.019 | -0.2807 | No |
| 89 | RAB27A | NA | 40682 | -0.020 | -0.2813 | No |
| 90 | LCE3C | NA | 40904 | -0.023 | -0.2847 | No |
| 91 | CD59 | NA | 40953 | -0.023 | -0.2848 | No |
| 92 | IL12RB1 | NA | 41014 | -0.024 | -0.2852 | No |
| 93 | CFHR2 | NA | 41420 | -0.029 | -0.2917 | No |
| 94 | AZU1 | NA | 41510 | -0.030 | -0.2924 | No |
| 95 | CLEC12B | NA | 41992 | -0.035 | -0.3002 | No |
| 96 | VAV1 | NA | 42188 | -0.038 | -0.3026 | No |
| 97 | RPS19 | NA | 42919 | -0.048 | -0.3145 | No |
| 98 | NOS2 | NA | 43066 | -0.049 | -0.3156 | No |
| 99 | C3 | NA | 43139 | -0.050 | -0.3154 | No |
| 100 | CAMP | NA | 43154 | -0.050 | -0.3141 | No |
| 101 | CD5L | NA | 43689 | -0.058 | -0.3221 | No |
| 102 | DEFA1 | NA | 43802 | -0.059 | -0.3223 | No |
| 103 | CFHR5 | NA | 43894 | -0.061 | -0.3221 | No |
| 104 | KLRC1 | NA | 43992 | -0.062 | -0.3219 | No |
| 105 | F2 | NA | 44044 | -0.062 | -0.3209 | No |
| 106 | DEFB132 | NA | 44159 | -0.064 | -0.3210 | No |
| 107 | CD1D | NA | 44239 | -0.065 | -0.3204 | No |
| 108 | CLEC2A | NA | 44623 | -0.071 | -0.3252 | No |
| 109 | RAET1E | NA | 44805 | -0.073 | -0.3262 | No |
| 110 | ARRB2 | NA | 45025 | -0.077 | -0.3278 | No |
| 111 | LAG3 | NA | 45326 | -0.081 | -0.3308 | No |
| 112 | HLA-G | NA | 45353 | -0.081 | -0.3287 | No |
| 113 | CCL13 | NA | 45642 | -0.086 | -0.3313 | No |
| 114 | IL4 | NA | 45895 | -0.090 | -0.3332 | No |
| 115 | CEACAM1 | NA | 46340 | -0.097 | -0.3383 | No |
| 116 | ELANE | NA | 47085 | -0.109 | -0.3486 | No |
| 117 | HMGN2 | NA | 47393 | -0.114 | -0.3506 | No |
| 118 | ULBP3 | NA | 47487 | -0.116 | -0.3487 | No |
| 119 | PGLYRP3 | NA | 48275 | -0.131 | -0.3591 | No |
| 120 | TYROBP | NA | 48389 | -0.132 | -0.3570 | No |
| 121 | FCER2 | NA | 48744 | -0.139 | -0.3591 | No |
| 122 | GNLY | NA | 49036 | -0.145 | -0.3599 | Yes |
| 123 | KLRK1 | NA | 49043 | -0.146 | -0.3555 | Yes |
| 124 | IL21 | NA | 49506 | -0.155 | -0.3591 | Yes |
| 125 | DEFA1B | NA | 49796 | -0.162 | -0.3594 | Yes |
| 126 | HSP90AB1 | NA | 50052 | -0.168 | -0.3588 | Yes |
| 127 | GAPDH | NA | 50120 | -0.169 | -0.3547 | Yes |
| 128 | DNASE1L3 | NA | 50126 | -0.169 | -0.3495 | Yes |
| 129 | CTSC | NA | 50256 | -0.172 | -0.3465 | Yes |
| 130 | KLRD1 | NA | 50561 | -0.178 | -0.3465 | Yes |
| 131 | IL13 | NA | 50585 | -0.179 | -0.3413 | Yes |
| 132 | PRF1 | NA | 50616 | -0.179 | -0.3363 | Yes |
| 133 | KIR3DL1 | NA | 50903 | -0.187 | -0.3357 | Yes |
| 134 | RASGRP1 | NA | 51143 | -0.193 | -0.3340 | Yes |
| 135 | ROMO1 | NA | 51187 | -0.194 | -0.3288 | Yes |
| 136 | PTPRC | NA | 51361 | -0.198 | -0.3257 | Yes |
| 137 | RPL30 | NA | 51577 | -0.204 | -0.3233 | Yes |
| 138 | SERPINB9 | NA | 51773 | -0.209 | -0.3203 | Yes |
| 139 | SH2D1A | NA | 51946 | -0.213 | -0.3168 | Yes |
| 140 | HPRT1 | NA | 51964 | -0.214 | -0.3104 | Yes |
| 141 | RAET1G | NA | 52044 | -0.216 | -0.3051 | Yes |
| 142 | GZMM | NA | 52105 | -0.218 | -0.2994 | Yes |
| 143 | IL18RAP | NA | 52639 | -0.233 | -0.3019 | Yes |
| 144 | CD55 | NA | 52968 | -0.243 | -0.3003 | Yes |
| 145 | CTSH | NA | 53038 | -0.247 | -0.2938 | Yes |
| 146 | MICA | NA | 53128 | -0.251 | -0.2876 | Yes |
| 147 | CR1L | NA | 53184 | -0.254 | -0.2806 | Yes |
| 148 | PGLYRP1 | NA | 53264 | -0.258 | -0.2740 | Yes |
| 149 | DEFA4 | NA | 53312 | -0.262 | -0.2667 | Yes |
| 150 | NECTIN2 | NA | 53322 | -0.262 | -0.2586 | Yes |
| 151 | IFNG | NA | 53327 | -0.263 | -0.2505 | Yes |
| 152 | APOL1 | NA | 53412 | -0.268 | -0.2436 | Yes |
| 153 | RNF19B | NA | 53427 | -0.270 | -0.2354 | Yes |
| 154 | NCR3 | NA | 53635 | -0.286 | -0.2303 | Yes |
| 155 | GZMB | NA | 53679 | -0.290 | -0.2220 | Yes |
| 156 | ULBP2 | NA | 53851 | -0.306 | -0.2155 | Yes |
| 157 | HLA-H | NA | 53882 | -0.309 | -0.2064 | Yes |
| 158 | NCR1 | NA | 53943 | -0.317 | -0.1976 | Yes |
| 159 | ARG1 | NA | 54035 | -0.331 | -0.1889 | Yes |
| 160 | ICAM1 | NA | 54181 | -0.359 | -0.1803 | Yes |
| 161 | S100A12 | NA | 54225 | -0.369 | -0.1696 | Yes |
| 162 | ULBP1 | NA | 54305 | -0.394 | -0.1587 | Yes |
| 163 | DEFA3 | NA | 54336 | -0.406 | -0.1465 | Yes |
| 164 | HLA-A | NA | 54414 | -0.439 | -0.1342 | Yes |
| 165 | PVR | NA | 54417 | -0.441 | -0.1204 | Yes |
| 166 | HLA-DRA | NA | 54422 | -0.444 | -0.1066 | Yes |
| 167 | B2M | NA | 54426 | -0.445 | -0.0927 | Yes |
| 168 | PF4 | NA | 54448 | -0.463 | -0.0786 | Yes |
| 169 | HLA-F | NA | 54455 | -0.467 | -0.0641 | Yes |
| 170 | KIR2DL4 | NA | 54465 | -0.472 | -0.0494 | Yes |
| 171 | HLA-DRB1 | NA | 54537 | -0.541 | -0.0338 | Yes |
| 172 | HLA-E | NA | 54547 | -0.553 | -0.0167 | Yes |
| 173 | HLA-B | NA | 54552 | -0.557 | 0.0007 | Yes |