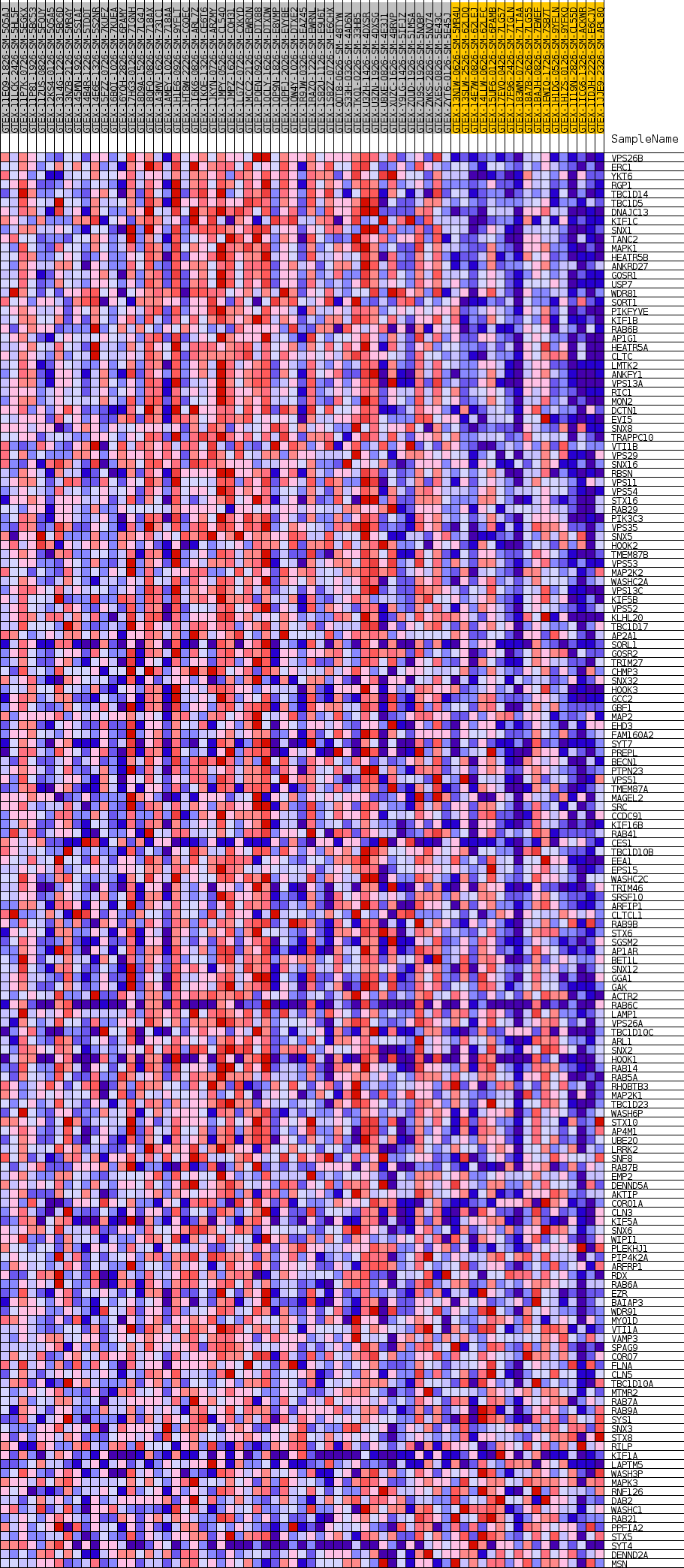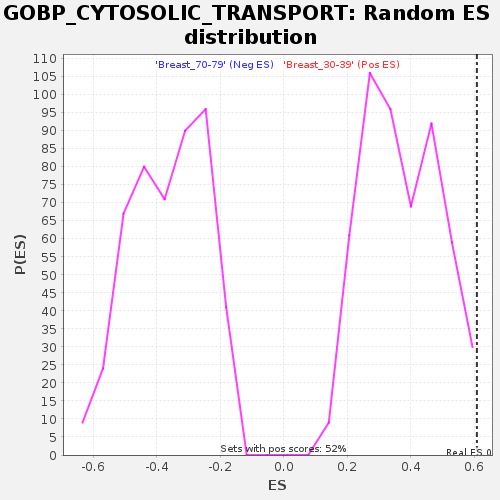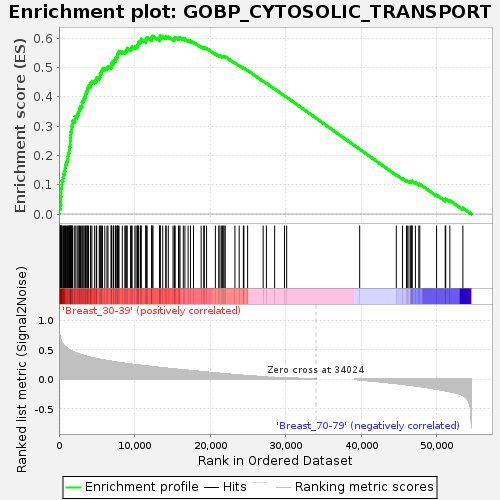
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_CYTOSOLIC_TRANSPORT |
| Enrichment Score (ES) | 0.607674 |
| Normalized Enrichment Score (NES) | 1.6357238 |
| Nominal p-value | 0.007662835 |
| FDR q-value | 0.24142395 |
| FWER p-Value | 0.834 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | VPS26B | NA | 11 | 0.876 | 0.0195 | Yes |
| 2 | ERC1 | NA | 202 | 0.672 | 0.0312 | Yes |
| 3 | YKT6 | NA | 257 | 0.652 | 0.0449 | Yes |
| 4 | RGP1 | NA | 259 | 0.652 | 0.0595 | Yes |
| 5 | TBC1D14 | NA | 297 | 0.639 | 0.0732 | Yes |
| 6 | TBC1D5 | NA | 299 | 0.638 | 0.0876 | Yes |
| 7 | DNAJC13 | NA | 373 | 0.619 | 0.1002 | Yes |
| 8 | KIF1C | NA | 391 | 0.615 | 0.1137 | Yes |
| 9 | SNX1 | NA | 530 | 0.588 | 0.1245 | Yes |
| 10 | TANC2 | NA | 608 | 0.575 | 0.1360 | Yes |
| 11 | MAPK1 | NA | 738 | 0.558 | 0.1462 | Yes |
| 12 | HEATR5B | NA | 806 | 0.550 | 0.1573 | Yes |
| 13 | ANKRD27 | NA | 865 | 0.544 | 0.1685 | Yes |
| 14 | GOSR1 | NA | 981 | 0.532 | 0.1784 | Yes |
| 15 | USP7 | NA | 1111 | 0.520 | 0.1878 | Yes |
| 16 | WDR81 | NA | 1174 | 0.514 | 0.1982 | Yes |
| 17 | SORT1 | NA | 1238 | 0.509 | 0.2085 | Yes |
| 18 | PIKFYVE | NA | 1353 | 0.499 | 0.2176 | Yes |
| 19 | KIF1B | NA | 1361 | 0.498 | 0.2287 | Yes |
| 20 | RAB6B | NA | 1476 | 0.490 | 0.2377 | Yes |
| 21 | AP1G1 | NA | 1483 | 0.489 | 0.2486 | Yes |
| 22 | HEATR5A | NA | 1502 | 0.488 | 0.2592 | Yes |
| 23 | CLTC | NA | 1540 | 0.486 | 0.2695 | Yes |
| 24 | LMTK2 | NA | 1569 | 0.484 | 0.2799 | Yes |
| 25 | ANKFY1 | NA | 1678 | 0.477 | 0.2886 | Yes |
| 26 | VPS13A | NA | 1683 | 0.477 | 0.2993 | Yes |
| 27 | RIC1 | NA | 1782 | 0.469 | 0.3081 | Yes |
| 28 | MON2 | NA | 1827 | 0.466 | 0.3177 | Yes |
| 29 | DCTN1 | NA | 2112 | 0.450 | 0.3227 | Yes |
| 30 | EVI5 | NA | 2115 | 0.450 | 0.3328 | Yes |
| 31 | SNX8 | NA | 2387 | 0.436 | 0.3376 | Yes |
| 32 | TRAPPC10 | NA | 2516 | 0.430 | 0.3450 | Yes |
| 33 | VTI1B | NA | 2653 | 0.424 | 0.3520 | Yes |
| 34 | VPS29 | NA | 2699 | 0.422 | 0.3607 | Yes |
| 35 | SNX16 | NA | 2839 | 0.416 | 0.3675 | Yes |
| 36 | RBSN | NA | 3041 | 0.408 | 0.3730 | Yes |
| 37 | VPS11 | NA | 3042 | 0.408 | 0.3822 | Yes |
| 38 | VPS54 | NA | 3178 | 0.403 | 0.3888 | Yes |
| 39 | STX16 | NA | 3308 | 0.398 | 0.3954 | Yes |
| 40 | RAB29 | NA | 3442 | 0.393 | 0.4018 | Yes |
| 41 | PIK3C3 | NA | 3507 | 0.391 | 0.4094 | Yes |
| 42 | VPS35 | NA | 3595 | 0.388 | 0.4165 | Yes |
| 43 | SNX5 | NA | 3720 | 0.383 | 0.4229 | Yes |
| 44 | HOOK2 | NA | 3802 | 0.380 | 0.4300 | Yes |
| 45 | TMEM87B | NA | 3902 | 0.377 | 0.4366 | Yes |
| 46 | VPS53 | NA | 4159 | 0.369 | 0.4402 | Yes |
| 47 | MAP2K2 | NA | 4205 | 0.368 | 0.4477 | Yes |
| 48 | WASHC2A | NA | 4360 | 0.363 | 0.4531 | Yes |
| 49 | VPS13C | NA | 4714 | 0.353 | 0.4545 | Yes |
| 50 | KIF5B | NA | 4934 | 0.347 | 0.4583 | Yes |
| 51 | VPS52 | NA | 4991 | 0.346 | 0.4651 | Yes |
| 52 | KLHL20 | NA | 5331 | 0.336 | 0.4664 | Yes |
| 53 | TBC1D17 | NA | 5431 | 0.334 | 0.4721 | Yes |
| 54 | AP2A1 | NA | 5439 | 0.334 | 0.4795 | Yes |
| 55 | SORL1 | NA | 5570 | 0.331 | 0.4846 | Yes |
| 56 | GOSR2 | NA | 5659 | 0.329 | 0.4903 | Yes |
| 57 | TRIM27 | NA | 5760 | 0.326 | 0.4959 | Yes |
| 58 | CHMP3 | NA | 6062 | 0.319 | 0.4975 | Yes |
| 59 | SNX32 | NA | 6351 | 0.313 | 0.4993 | Yes |
| 60 | HOOK3 | NA | 6481 | 0.310 | 0.5039 | Yes |
| 61 | GCC2 | NA | 6886 | 0.301 | 0.5032 | Yes |
| 62 | GBF1 | NA | 6966 | 0.300 | 0.5085 | Yes |
| 63 | MAP2 | NA | 6975 | 0.299 | 0.5151 | Yes |
| 64 | EHD3 | NA | 7201 | 0.295 | 0.5176 | Yes |
| 65 | FAM160A2 | NA | 7241 | 0.294 | 0.5236 | Yes |
| 66 | SYT7 | NA | 7477 | 0.289 | 0.5258 | Yes |
| 67 | PREPL | NA | 7490 | 0.289 | 0.5321 | Yes |
| 68 | BECN1 | NA | 7659 | 0.286 | 0.5354 | Yes |
| 69 | PTPN23 | NA | 7730 | 0.284 | 0.5405 | Yes |
| 70 | VPS51 | NA | 7737 | 0.284 | 0.5468 | Yes |
| 71 | TMEM87A | NA | 7858 | 0.282 | 0.5510 | Yes |
| 72 | MAGEL2 | NA | 7945 | 0.280 | 0.5557 | Yes |
| 73 | SRC | NA | 8379 | 0.272 | 0.5539 | Yes |
| 74 | CCDC91 | NA | 8717 | 0.266 | 0.5537 | Yes |
| 75 | KIF16B | NA | 8829 | 0.264 | 0.5576 | Yes |
| 76 | RAB41 | NA | 8966 | 0.262 | 0.5610 | Yes |
| 77 | CES1 | NA | 9021 | 0.261 | 0.5659 | Yes |
| 78 | TBC1D10B | NA | 9475 | 0.253 | 0.5633 | Yes |
| 79 | EEA1 | NA | 9609 | 0.251 | 0.5665 | Yes |
| 80 | EPS15 | NA | 9672 | 0.250 | 0.5709 | Yes |
| 81 | WASHC2C | NA | 10047 | 0.244 | 0.5696 | Yes |
| 82 | TRIM46 | NA | 10211 | 0.241 | 0.5720 | Yes |
| 83 | SRSF10 | NA | 10397 | 0.238 | 0.5740 | Yes |
| 84 | ARFIP1 | NA | 10422 | 0.238 | 0.5789 | Yes |
| 85 | CLTCL1 | NA | 10491 | 0.237 | 0.5830 | Yes |
| 86 | RAB9B | NA | 10560 | 0.236 | 0.5870 | Yes |
| 87 | STX6 | NA | 10790 | 0.232 | 0.5880 | Yes |
| 88 | SGSM2 | NA | 10856 | 0.231 | 0.5920 | Yes |
| 89 | AP1AR | NA | 10871 | 0.231 | 0.5970 | Yes |
| 90 | BET1L | NA | 11486 | 0.222 | 0.5907 | Yes |
| 91 | SNX12 | NA | 11531 | 0.221 | 0.5949 | Yes |
| 92 | GGA1 | NA | 11540 | 0.221 | 0.5997 | Yes |
| 93 | GAK | NA | 11664 | 0.219 | 0.6024 | Yes |
| 94 | ACTR2 | NA | 12288 | 0.211 | 0.5957 | Yes |
| 95 | RAB6C | NA | 12293 | 0.211 | 0.6004 | Yes |
| 96 | LAMP1 | NA | 12298 | 0.211 | 0.6050 | Yes |
| 97 | VPS26A | NA | 12442 | 0.209 | 0.6071 | Yes |
| 98 | TBC1D10C | NA | 13328 | 0.196 | 0.5953 | Yes |
| 99 | ARL1 | NA | 13370 | 0.195 | 0.5989 | Yes |
| 100 | SNX2 | NA | 13371 | 0.195 | 0.6033 | Yes |
| 101 | HOOK1 | NA | 13374 | 0.195 | 0.6077 | Yes |
| 102 | RAB14 | NA | 13732 | 0.190 | 0.6054 | No |
| 103 | RAB5A | NA | 14128 | 0.185 | 0.6023 | No |
| 104 | RHOBTB3 | NA | 14184 | 0.184 | 0.6054 | No |
| 105 | MAP2K1 | NA | 14468 | 0.181 | 0.6043 | No |
| 106 | TBC1D23 | NA | 15095 | 0.173 | 0.5967 | No |
| 107 | WASH6P | NA | 15266 | 0.171 | 0.5974 | No |
| 108 | STX10 | NA | 15295 | 0.170 | 0.6007 | No |
| 109 | AP4M1 | NA | 15375 | 0.169 | 0.6031 | No |
| 110 | UBE2O | NA | 15816 | 0.164 | 0.5987 | No |
| 111 | LRRK2 | NA | 15895 | 0.163 | 0.6009 | No |
| 112 | SNF8 | NA | 16020 | 0.161 | 0.6023 | No |
| 113 | RAB7B | NA | 16457 | 0.156 | 0.5978 | No |
| 114 | EMP2 | NA | 16669 | 0.153 | 0.5973 | No |
| 115 | DENND5A | NA | 17092 | 0.148 | 0.5929 | No |
| 116 | AKTIP | NA | 17403 | 0.144 | 0.5905 | No |
| 117 | CORO1A | NA | 17805 | 0.139 | 0.5862 | No |
| 118 | CLN3 | NA | 18819 | 0.126 | 0.5705 | No |
| 119 | KIF5A | NA | 19161 | 0.122 | 0.5670 | No |
| 120 | SNX6 | NA | 19235 | 0.121 | 0.5683 | No |
| 121 | WIPI1 | NA | 19536 | 0.117 | 0.5655 | No |
| 122 | PLEKHJ1 | NA | 20699 | 0.103 | 0.5464 | No |
| 123 | PIP4K2A | NA | 21146 | 0.098 | 0.5405 | No |
| 124 | ARFRP1 | NA | 21309 | 0.096 | 0.5396 | No |
| 125 | RDX | NA | 21542 | 0.093 | 0.5375 | No |
| 126 | RAB6A | NA | 21622 | 0.092 | 0.5381 | No |
| 127 | EZR | NA | 21731 | 0.091 | 0.5382 | No |
| 128 | BAIAP3 | NA | 21858 | 0.090 | 0.5379 | No |
| 129 | WDR91 | NA | 22006 | 0.088 | 0.5372 | No |
| 130 | MYO1D | NA | 23288 | 0.074 | 0.5153 | No |
| 131 | VTI1A | NA | 23852 | 0.068 | 0.5065 | No |
| 132 | VAMP3 | NA | 24404 | 0.063 | 0.4978 | No |
| 133 | SPAG9 | NA | 24476 | 0.062 | 0.4979 | No |
| 134 | CORO7 | NA | 24969 | 0.058 | 0.4902 | No |
| 135 | FLNA | NA | 27028 | 0.039 | 0.4532 | No |
| 136 | CLN5 | NA | 27457 | 0.036 | 0.4462 | No |
| 137 | TBC1D10A | NA | 28547 | 0.029 | 0.4268 | No |
| 138 | MTMR2 | NA | 29851 | 0.021 | 0.4034 | No |
| 139 | RAB7A | NA | 30138 | 0.020 | 0.3986 | No |
| 140 | RAB9A | NA | 39802 | -0.009 | 0.2212 | No |
| 141 | SYS1 | NA | 44652 | -0.071 | 0.1338 | No |
| 142 | SNX3 | NA | 45467 | -0.083 | 0.1207 | No |
| 143 | STX8 | NA | 46000 | -0.091 | 0.1130 | No |
| 144 | RILP | NA | 46105 | -0.093 | 0.1132 | No |
| 145 | KIF1A | NA | 46327 | -0.096 | 0.1113 | No |
| 146 | LAPTM5 | NA | 46576 | -0.100 | 0.1090 | No |
| 147 | WASH3P | NA | 46600 | -0.101 | 0.1108 | No |
| 148 | MAPK3 | NA | 46700 | -0.102 | 0.1113 | No |
| 149 | RNF126 | NA | 46754 | -0.103 | 0.1126 | No |
| 150 | DAB2 | NA | 47181 | -0.111 | 0.1073 | No |
| 151 | WASHC1 | NA | 47632 | -0.119 | 0.1017 | No |
| 152 | RAB21 | NA | 47759 | -0.121 | 0.1021 | No |
| 153 | PPFIA2 | NA | 49983 | -0.166 | 0.0650 | No |
| 154 | STX5 | NA | 51123 | -0.192 | 0.0484 | No |
| 155 | SYT4 | NA | 51197 | -0.194 | 0.0515 | No |
| 156 | DENND2A | NA | 51741 | -0.208 | 0.0462 | No |
| 157 | MSN | NA | 53465 | -0.273 | 0.0207 | No |