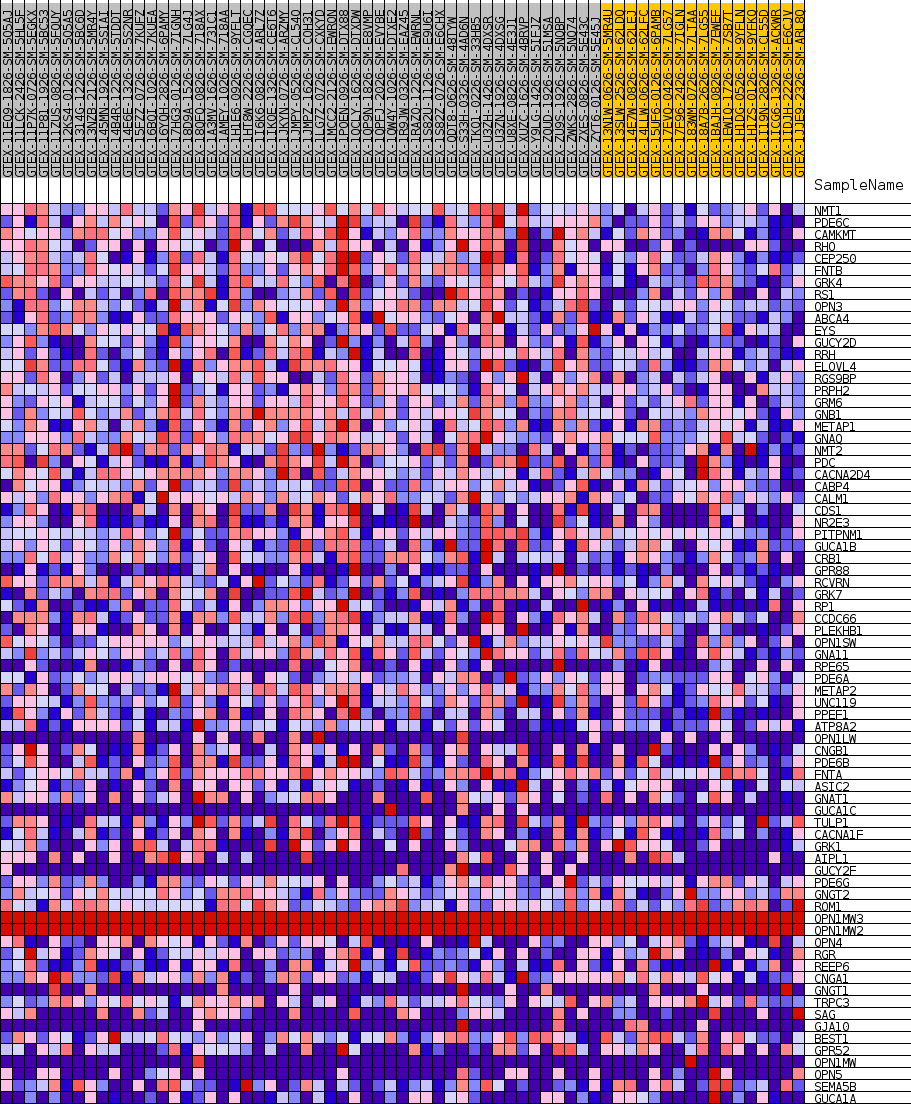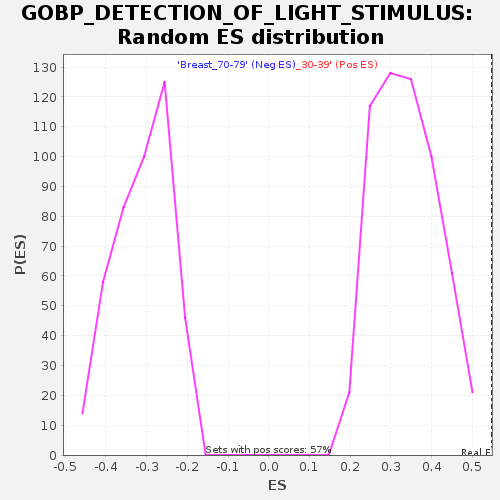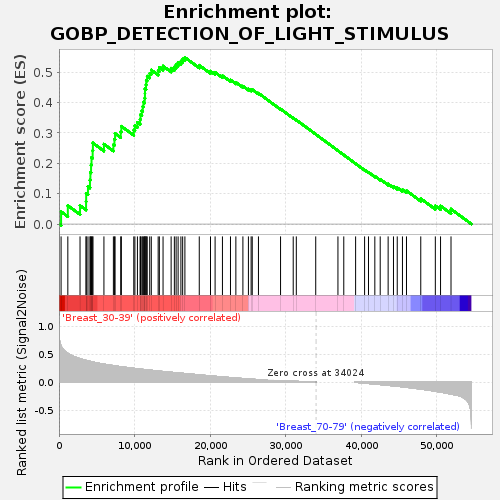
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_DETECTION_OF_LIGHT_STIMULUS |
| Enrichment Score (ES) | 0.54828525 |
| Normalized Enrichment Score (NES) | 1.6278166 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.2354168 |
| FWER p-Value | 0.852 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | NMT1 | NA | 276 | 0.648 | 0.0408 | Yes |
| 2 | PDE6C | NA | 1169 | 0.514 | 0.0609 | Yes |
| 3 | CAMKMT | NA | 2784 | 0.418 | 0.0609 | Yes |
| 4 | RHO | NA | 3584 | 0.388 | 0.0737 | Yes |
| 5 | CEP250 | NA | 3598 | 0.388 | 0.1010 | Yes |
| 6 | FNTB | NA | 3834 | 0.379 | 0.1235 | Yes |
| 7 | GRK4 | NA | 4100 | 0.371 | 0.1449 | Yes |
| 8 | RS1 | NA | 4154 | 0.370 | 0.1701 | Yes |
| 9 | OPN3 | NA | 4244 | 0.367 | 0.1945 | Yes |
| 10 | ABCA4 | NA | 4305 | 0.365 | 0.2192 | Yes |
| 11 | EYS | NA | 4456 | 0.361 | 0.2420 | Yes |
| 12 | GUCY2D | NA | 4487 | 0.360 | 0.2670 | Yes |
| 13 | RRH | NA | 5947 | 0.322 | 0.2630 | Yes |
| 14 | ELOVL4 | NA | 7199 | 0.295 | 0.2610 | Yes |
| 15 | RGS9BP | NA | 7338 | 0.292 | 0.2791 | Yes |
| 16 | PRPH2 | NA | 7423 | 0.290 | 0.2982 | Yes |
| 17 | GRM6 | NA | 8170 | 0.276 | 0.3040 | Yes |
| 18 | GNB1 | NA | 8256 | 0.274 | 0.3219 | Yes |
| 19 | METAP1 | NA | 9900 | 0.246 | 0.3092 | Yes |
| 20 | GNAQ | NA | 10046 | 0.244 | 0.3238 | Yes |
| 21 | NMT2 | NA | 10381 | 0.238 | 0.3345 | Yes |
| 22 | PDC | NA | 10735 | 0.233 | 0.3445 | Yes |
| 23 | CACNA2D4 | NA | 10786 | 0.232 | 0.3601 | Yes |
| 24 | CABP4 | NA | 10966 | 0.229 | 0.3730 | Yes |
| 25 | CALM1 | NA | 11085 | 0.228 | 0.3870 | Yes |
| 26 | CDS1 | NA | 11169 | 0.227 | 0.4015 | Yes |
| 27 | NR2E3 | NA | 11348 | 0.224 | 0.4141 | Yes |
| 28 | PITPNM1 | NA | 11378 | 0.223 | 0.4294 | Yes |
| 29 | GUCA1B | NA | 11382 | 0.223 | 0.4452 | Yes |
| 30 | CRB1 | NA | 11520 | 0.221 | 0.4583 | Yes |
| 31 | GPR88 | NA | 11550 | 0.221 | 0.4734 | Yes |
| 32 | RCVRN | NA | 11678 | 0.219 | 0.4866 | Yes |
| 33 | GRK7 | NA | 12009 | 0.214 | 0.4958 | Yes |
| 34 | RP1 | NA | 12218 | 0.212 | 0.5069 | Yes |
| 35 | CCDC66 | NA | 13127 | 0.199 | 0.5044 | Yes |
| 36 | PLEKHB1 | NA | 13286 | 0.197 | 0.5154 | Yes |
| 37 | OPN1SW | NA | 13774 | 0.190 | 0.5199 | Yes |
| 38 | GNA11 | NA | 14843 | 0.176 | 0.5128 | Yes |
| 39 | RPE65 | NA | 15271 | 0.170 | 0.5170 | Yes |
| 40 | PDE6A | NA | 15502 | 0.168 | 0.5247 | Yes |
| 41 | METAP2 | NA | 15768 | 0.164 | 0.5314 | Yes |
| 42 | UNC119 | NA | 16137 | 0.160 | 0.5360 | Yes |
| 43 | PPEF1 | NA | 16370 | 0.157 | 0.5428 | Yes |
| 44 | ATP8A2 | NA | 16666 | 0.153 | 0.5483 | Yes |
| 45 | OPN1LW | NA | 18568 | 0.129 | 0.5226 | No |
| 46 | CNGB1 | NA | 20056 | 0.111 | 0.5031 | No |
| 47 | PDE6B | NA | 20664 | 0.104 | 0.4993 | No |
| 48 | FNTA | NA | 21636 | 0.092 | 0.4880 | No |
| 49 | ASIC2 | NA | 22701 | 0.080 | 0.4742 | No |
| 50 | GNAT1 | NA | 23413 | 0.073 | 0.4663 | No |
| 51 | GUCA1C | NA | 24344 | 0.064 | 0.4538 | No |
| 52 | TULP1 | NA | 25074 | 0.057 | 0.4444 | No |
| 53 | CACNA1F | NA | 25433 | 0.053 | 0.4416 | No |
| 54 | GRK1 | NA | 25581 | 0.052 | 0.4426 | No |
| 55 | AIPL1 | NA | 26403 | 0.044 | 0.4306 | No |
| 56 | GUCY2F | NA | 29329 | 0.024 | 0.3787 | No |
| 57 | PDE6G | NA | 31005 | 0.015 | 0.3490 | No |
| 58 | GNGT2 | NA | 31415 | 0.013 | 0.3425 | No |
| 59 | ROM1 | NA | 33985 | 0.001 | 0.2954 | No |
| 60 | OPN1MW3 | NA | 36926 | 0.000 | 0.2415 | No |
| 61 | OPN1MW2 | NA | 37698 | 0.000 | 0.2273 | No |
| 62 | OPN4 | NA | 39266 | -0.002 | 0.1987 | No |
| 63 | RGR | NA | 40455 | -0.017 | 0.1781 | No |
| 64 | REEP6 | NA | 40970 | -0.024 | 0.1704 | No |
| 65 | CNGA1 | NA | 41809 | -0.033 | 0.1573 | No |
| 66 | GNGT1 | NA | 42521 | -0.042 | 0.1473 | No |
| 67 | TRPC3 | NA | 43571 | -0.056 | 0.1320 | No |
| 68 | SAG | NA | 44278 | -0.066 | 0.1237 | No |
| 69 | GJA10 | NA | 44776 | -0.073 | 0.1198 | No |
| 70 | BEST1 | NA | 45466 | -0.083 | 0.1130 | No |
| 71 | GPR52 | NA | 45999 | -0.091 | 0.1097 | No |
| 72 | OPN1MW | NA | 47890 | -0.124 | 0.0838 | No |
| 73 | OPN5 | NA | 49818 | -0.162 | 0.0600 | No |
| 74 | SEMA5B | NA | 50509 | -0.177 | 0.0598 | No |
| 75 | GUCA1A | NA | 51894 | -0.212 | 0.0495 | No |