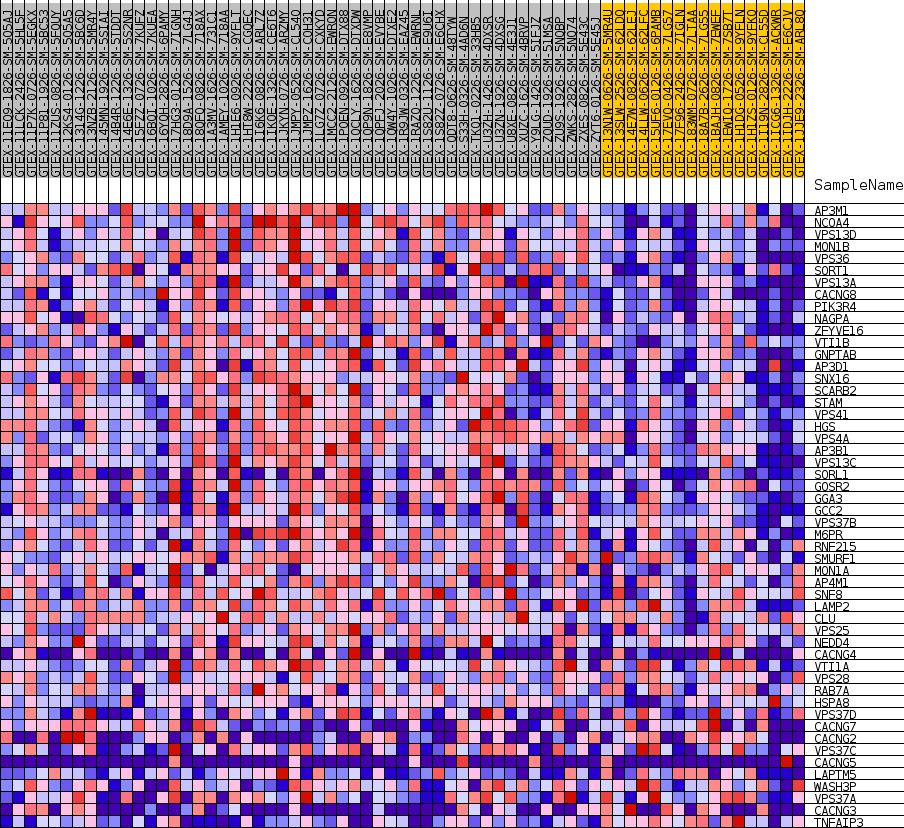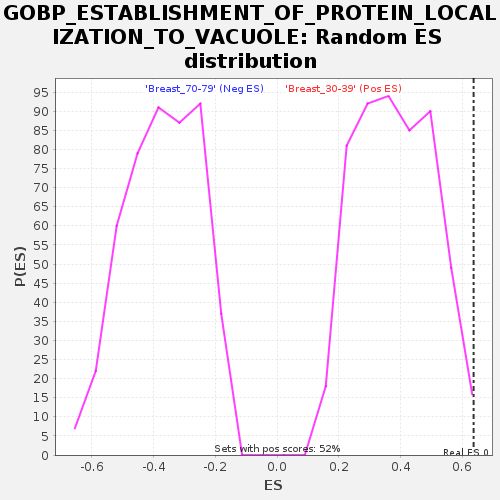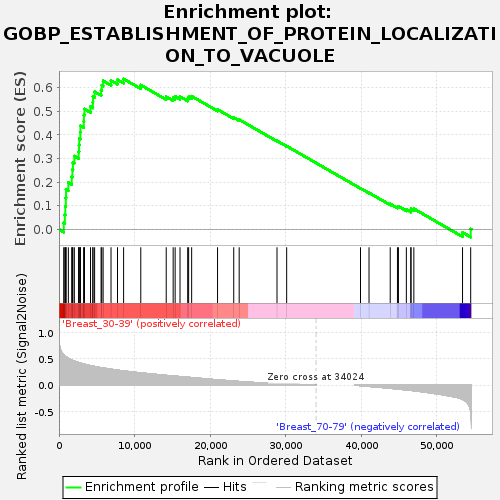
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_ESTABLISHMENT_OF_PROTEIN_LOCALIZATION_TO_VACUOLE |
| Enrichment Score (ES) | 0.6364749 |
| Normalized Enrichment Score (NES) | 1.6617539 |
| Nominal p-value | 0.0038095238 |
| FDR q-value | 0.23918153 |
| FWER p-Value | 0.769 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | AP3M1 | NA | 623 | 0.573 | 0.0274 | Yes |
| 2 | NCOA4 | NA | 773 | 0.553 | 0.0621 | Yes |
| 3 | VPS13D | NA | 831 | 0.548 | 0.0982 | Yes |
| 4 | MON1B | NA | 898 | 0.542 | 0.1336 | Yes |
| 5 | VPS36 | NA | 937 | 0.536 | 0.1693 | Yes |
| 6 | SORT1 | NA | 1238 | 0.509 | 0.1982 | Yes |
| 7 | VPS13A | NA | 1683 | 0.477 | 0.2224 | Yes |
| 8 | CACNG8 | NA | 1776 | 0.469 | 0.2525 | Yes |
| 9 | PIK3R4 | NA | 1840 | 0.466 | 0.2828 | Yes |
| 10 | NAGPA | NA | 2030 | 0.454 | 0.3101 | Yes |
| 11 | ZFYVE16 | NA | 2596 | 0.427 | 0.3287 | Yes |
| 12 | VTI1B | NA | 2653 | 0.424 | 0.3564 | Yes |
| 13 | GNPTAB | NA | 2681 | 0.423 | 0.3846 | Yes |
| 14 | AP3D1 | NA | 2825 | 0.416 | 0.4102 | Yes |
| 15 | SNX16 | NA | 2839 | 0.416 | 0.4381 | Yes |
| 16 | SCARB2 | NA | 3276 | 0.399 | 0.4571 | Yes |
| 17 | STAM | NA | 3298 | 0.398 | 0.4837 | Yes |
| 18 | VPS41 | NA | 3371 | 0.396 | 0.5092 | Yes |
| 19 | HGS | NA | 4170 | 0.369 | 0.5195 | Yes |
| 20 | VPS4A | NA | 4481 | 0.360 | 0.5382 | Yes |
| 21 | AP3B1 | NA | 4493 | 0.360 | 0.5624 | Yes |
| 22 | VPS13C | NA | 4714 | 0.353 | 0.5823 | Yes |
| 23 | SORL1 | NA | 5570 | 0.331 | 0.5890 | Yes |
| 24 | GOSR2 | NA | 5659 | 0.329 | 0.6096 | Yes |
| 25 | GGA3 | NA | 5841 | 0.324 | 0.6283 | Yes |
| 26 | GCC2 | NA | 6886 | 0.301 | 0.6295 | Yes |
| 27 | VPS37B | NA | 7744 | 0.284 | 0.6331 | Yes |
| 28 | M6PR | NA | 8553 | 0.269 | 0.6365 | Yes |
| 29 | RNF215 | NA | 10826 | 0.231 | 0.6105 | No |
| 30 | SMURF1 | NA | 14194 | 0.184 | 0.5612 | No |
| 31 | MON1A | NA | 15112 | 0.172 | 0.5561 | No |
| 32 | AP4M1 | NA | 15375 | 0.169 | 0.5627 | No |
| 33 | SNF8 | NA | 16020 | 0.161 | 0.5618 | No |
| 34 | LAMP2 | NA | 17036 | 0.149 | 0.5533 | No |
| 35 | CLU | NA | 17160 | 0.147 | 0.5610 | No |
| 36 | VPS25 | NA | 17561 | 0.142 | 0.5633 | No |
| 37 | NEDD4 | NA | 20989 | 0.100 | 0.5072 | No |
| 38 | CACNG4 | NA | 23131 | 0.076 | 0.4731 | No |
| 39 | VTI1A | NA | 23852 | 0.068 | 0.4645 | No |
| 40 | VPS28 | NA | 28859 | 0.027 | 0.3746 | No |
| 41 | RAB7A | NA | 30138 | 0.020 | 0.3525 | No |
| 42 | HSPA8 | NA | 39913 | -0.011 | 0.1740 | No |
| 43 | VPS37D | NA | 41041 | -0.024 | 0.1550 | No |
| 44 | CACNG7 | NA | 43846 | -0.060 | 0.1076 | No |
| 45 | CACNG2 | NA | 44804 | -0.073 | 0.0950 | No |
| 46 | VPS37C | NA | 44935 | -0.075 | 0.0977 | No |
| 47 | CACNG5 | NA | 45981 | -0.091 | 0.0847 | No |
| 48 | LAPTM5 | NA | 46576 | -0.100 | 0.0807 | No |
| 49 | WASH3P | NA | 46600 | -0.101 | 0.0871 | No |
| 50 | VPS37A | NA | 46974 | -0.107 | 0.0875 | No |
| 51 | CACNG3 | NA | 53420 | -0.269 | -0.0125 | No |
| 52 | TNFAIP3 | NA | 54500 | -0.501 | 0.0017 | No |