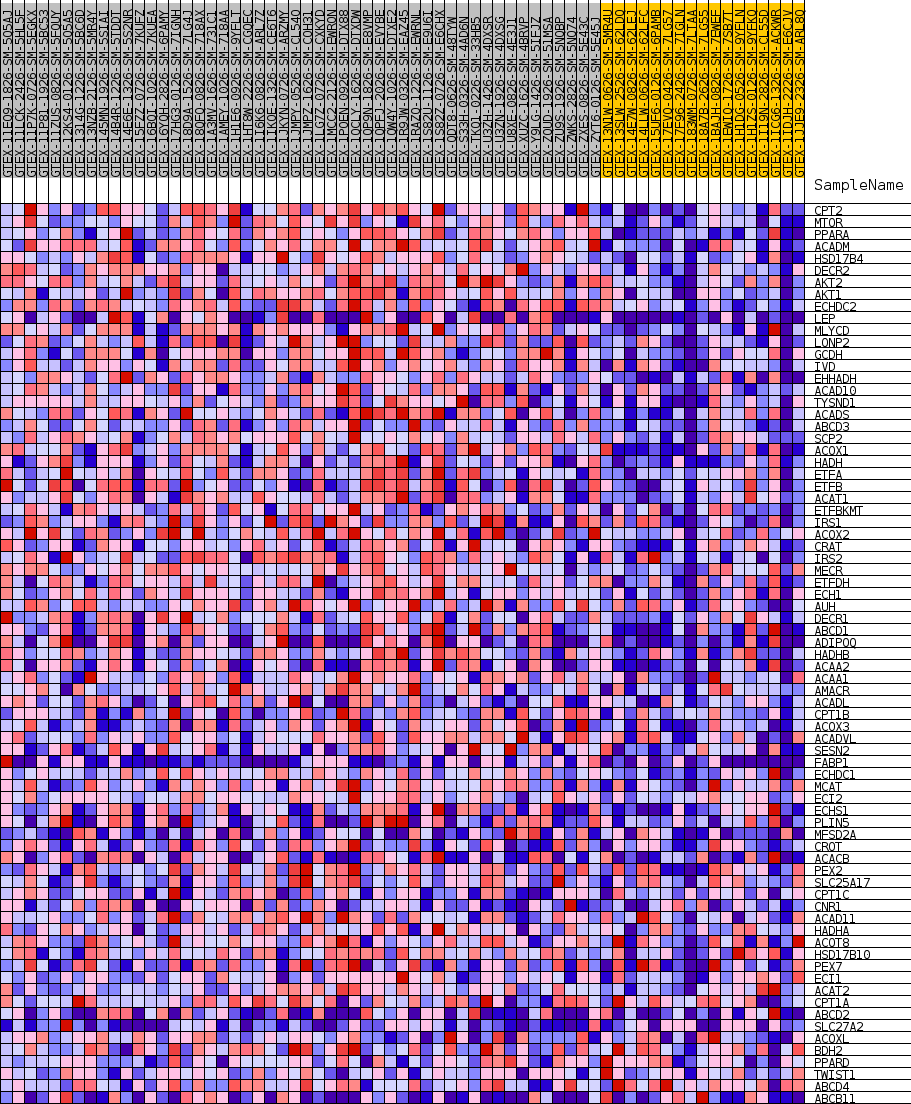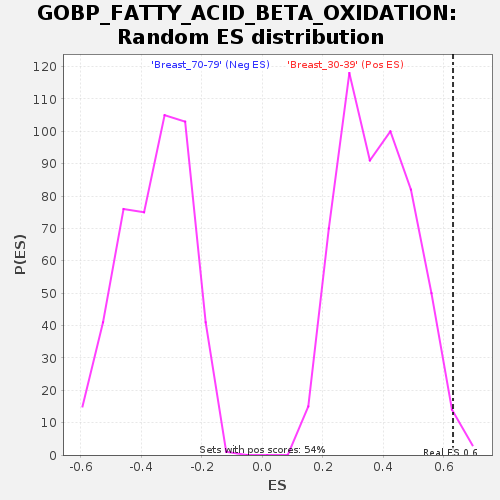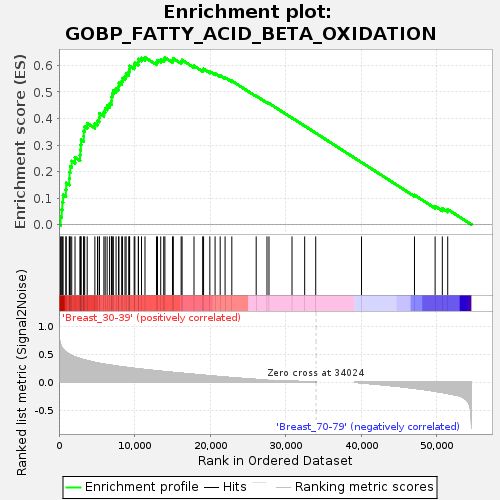
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_FATTY_ACID_BETA_OXIDATION |
| Enrichment Score (ES) | 0.6303864 |
| Normalized Enrichment Score (NES) | 1.6710024 |
| Nominal p-value | 0.009208103 |
| FDR q-value | 0.2685075 |
| FWER p-Value | 0.751 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | CPT2 | NA | 210 | 0.668 | 0.0288 | Yes |
| 2 | MTOR | NA | 355 | 0.623 | 0.0567 | Yes |
| 3 | PPARA | NA | 438 | 0.605 | 0.0848 | Yes |
| 4 | ACADM | NA | 542 | 0.586 | 0.1116 | Yes |
| 5 | HSD17B4 | NA | 909 | 0.540 | 0.1314 | Yes |
| 6 | DECR2 | NA | 954 | 0.535 | 0.1567 | Yes |
| 7 | AKT2 | NA | 1358 | 0.498 | 0.1737 | Yes |
| 8 | AKT1 | NA | 1390 | 0.496 | 0.1974 | Yes |
| 9 | ECHDC2 | NA | 1496 | 0.489 | 0.2194 | Yes |
| 10 | LEP | NA | 1684 | 0.477 | 0.2393 | Yes |
| 11 | MLYCD | NA | 2121 | 0.450 | 0.2533 | Yes |
| 12 | LONP2 | NA | 2778 | 0.419 | 0.2618 | Yes |
| 13 | GCDH | NA | 2826 | 0.416 | 0.2813 | Yes |
| 14 | IVD | NA | 2878 | 0.414 | 0.3006 | Yes |
| 15 | EHHADH | NA | 2929 | 0.412 | 0.3199 | Yes |
| 16 | ACAD10 | NA | 3267 | 0.399 | 0.3332 | Yes |
| 17 | TYSND1 | NA | 3274 | 0.399 | 0.3527 | Yes |
| 18 | ACADS | NA | 3399 | 0.395 | 0.3697 | Yes |
| 19 | ABCD3 | NA | 3733 | 0.382 | 0.3823 | Yes |
| 20 | SCP2 | NA | 4749 | 0.352 | 0.3809 | Yes |
| 21 | ACOX1 | NA | 5098 | 0.343 | 0.3914 | Yes |
| 22 | HADH | NA | 5324 | 0.336 | 0.4037 | Yes |
| 23 | ETFA | NA | 5356 | 0.336 | 0.4195 | Yes |
| 24 | ETFB | NA | 5943 | 0.322 | 0.4246 | Yes |
| 25 | ACAT1 | NA | 6111 | 0.318 | 0.4371 | Yes |
| 26 | ETFBKMT | NA | 6372 | 0.312 | 0.4476 | Yes |
| 27 | IRS1 | NA | 6700 | 0.305 | 0.4565 | Yes |
| 28 | ACOX2 | NA | 6955 | 0.300 | 0.4665 | Yes |
| 29 | CRAT | NA | 6969 | 0.300 | 0.4810 | Yes |
| 30 | IRS2 | NA | 7052 | 0.298 | 0.4940 | Yes |
| 31 | MECR | NA | 7192 | 0.295 | 0.5060 | Yes |
| 32 | ETFDH | NA | 7555 | 0.288 | 0.5134 | Yes |
| 33 | ECH1 | NA | 7889 | 0.281 | 0.5211 | Yes |
| 34 | AUH | NA | 7930 | 0.280 | 0.5341 | Yes |
| 35 | DECR1 | NA | 8300 | 0.274 | 0.5407 | Yes |
| 36 | ABCD1 | NA | 8410 | 0.272 | 0.5520 | Yes |
| 37 | ADIPOQ | NA | 8737 | 0.266 | 0.5590 | Yes |
| 38 | HADHB | NA | 8884 | 0.263 | 0.5692 | Yes |
| 39 | ACAA2 | NA | 9231 | 0.257 | 0.5755 | Yes |
| 40 | ACAA1 | NA | 9324 | 0.255 | 0.5863 | Yes |
| 41 | AMACR | NA | 9352 | 0.255 | 0.5983 | Yes |
| 42 | ACADL | NA | 9951 | 0.245 | 0.5993 | Yes |
| 43 | CPT1B | NA | 10043 | 0.244 | 0.6096 | Yes |
| 44 | ACOX3 | NA | 10503 | 0.236 | 0.6127 | Yes |
| 45 | ACADVL | NA | 10516 | 0.236 | 0.6241 | Yes |
| 46 | SESN2 | NA | 10921 | 0.230 | 0.6279 | Yes |
| 47 | FABP1 | NA | 11383 | 0.223 | 0.6304 | Yes |
| 48 | ECHDC1 | NA | 12905 | 0.202 | 0.6124 | No |
| 49 | MCAT | NA | 13040 | 0.200 | 0.6197 | No |
| 50 | ECI2 | NA | 13465 | 0.194 | 0.6214 | No |
| 51 | ECHS1 | NA | 13864 | 0.189 | 0.6234 | No |
| 52 | PLIN5 | NA | 14018 | 0.186 | 0.6297 | No |
| 53 | MFSD2A | NA | 15023 | 0.173 | 0.6197 | No |
| 54 | CROT | NA | 15134 | 0.172 | 0.6262 | No |
| 55 | ACACB | NA | 16170 | 0.159 | 0.6150 | No |
| 56 | PEX2 | NA | 16293 | 0.158 | 0.6204 | No |
| 57 | SLC25A17 | NA | 17872 | 0.138 | 0.5983 | No |
| 58 | CPT1C | NA | 19017 | 0.124 | 0.5833 | No |
| 59 | CNR1 | NA | 19137 | 0.122 | 0.5871 | No |
| 60 | ACAD11 | NA | 19969 | 0.112 | 0.5773 | No |
| 61 | HADHA | NA | 20662 | 0.104 | 0.5697 | No |
| 62 | ACOT8 | NA | 21338 | 0.096 | 0.5620 | No |
| 63 | HSD17B10 | NA | 21987 | 0.088 | 0.5545 | No |
| 64 | PEX7 | NA | 22878 | 0.079 | 0.5420 | No |
| 65 | ECI1 | NA | 26103 | 0.047 | 0.4851 | No |
| 66 | ACAT2 | NA | 27532 | 0.036 | 0.4607 | No |
| 67 | CPT1A | NA | 27796 | 0.034 | 0.4575 | No |
| 68 | ABCD2 | NA | 30846 | 0.016 | 0.4024 | No |
| 69 | SLC27A2 | NA | 32521 | 0.009 | 0.3721 | No |
| 70 | ACOXL | NA | 33984 | 0.001 | 0.3453 | No |
| 71 | BDH2 | NA | 40047 | -0.012 | 0.2347 | No |
| 72 | PPARD | NA | 47058 | -0.108 | 0.1114 | No |
| 73 | TWIST1 | NA | 49789 | -0.162 | 0.0693 | No |
| 74 | ABCD4 | NA | 50744 | -0.183 | 0.0607 | No |
| 75 | ABCB11 | NA | 51458 | -0.201 | 0.0575 | No |