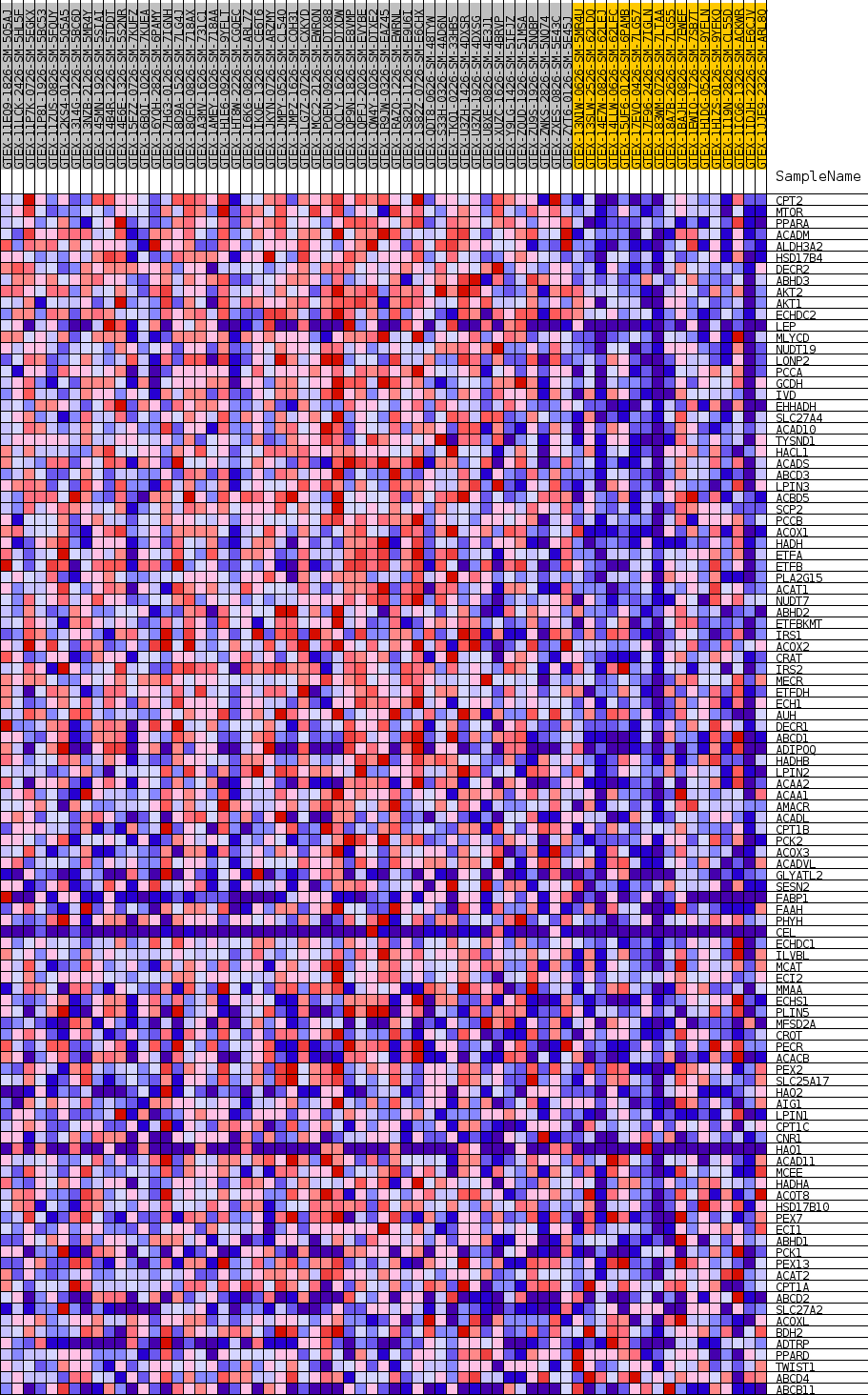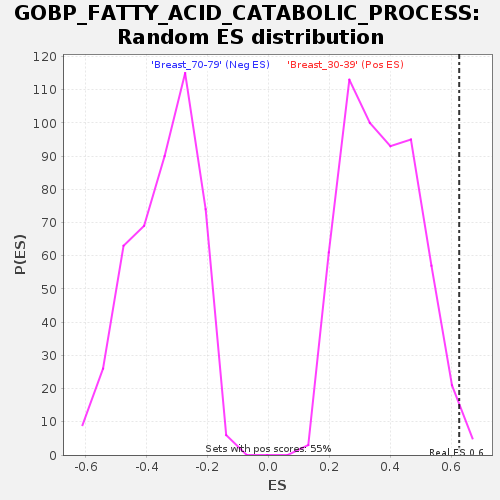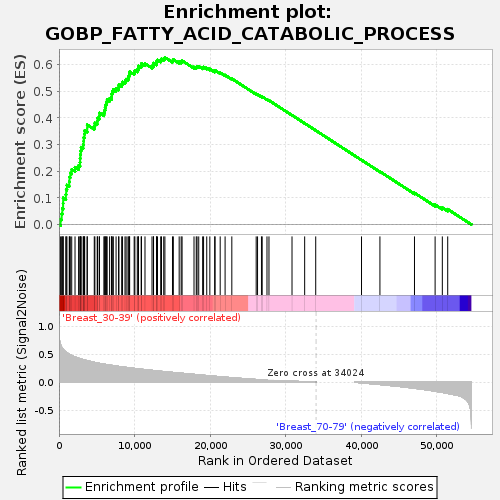
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_FATTY_ACID_CATABOLIC_PROCESS |
| Enrichment Score (ES) | 0.62649095 |
| Normalized Enrichment Score (NES) | 1.6797605 |
| Nominal p-value | 0.01459854 |
| FDR q-value | 0.27899468 |
| FWER p-Value | 0.72 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | CPT2 | NA | 210 | 0.668 | 0.0201 | Yes |
| 2 | MTOR | NA | 355 | 0.623 | 0.0398 | Yes |
| 3 | PPARA | NA | 438 | 0.605 | 0.0599 | Yes |
| 4 | ACADM | NA | 542 | 0.586 | 0.0791 | Yes |
| 5 | ALDH3A2 | NA | 550 | 0.585 | 0.0999 | Yes |
| 6 | HSD17B4 | NA | 909 | 0.540 | 0.1127 | Yes |
| 7 | DECR2 | NA | 954 | 0.535 | 0.1310 | Yes |
| 8 | ABHD3 | NA | 1049 | 0.525 | 0.1481 | Yes |
| 9 | AKT2 | NA | 1358 | 0.498 | 0.1603 | Yes |
| 10 | AKT1 | NA | 1390 | 0.496 | 0.1775 | Yes |
| 11 | ECHDC2 | NA | 1496 | 0.489 | 0.1931 | Yes |
| 12 | LEP | NA | 1684 | 0.477 | 0.2067 | Yes |
| 13 | MLYCD | NA | 2121 | 0.450 | 0.2149 | Yes |
| 14 | NUDT19 | NA | 2593 | 0.427 | 0.2215 | Yes |
| 15 | LONP2 | NA | 2778 | 0.419 | 0.2331 | Yes |
| 16 | PCCA | NA | 2794 | 0.418 | 0.2478 | Yes |
| 17 | GCDH | NA | 2826 | 0.416 | 0.2622 | Yes |
| 18 | IVD | NA | 2878 | 0.414 | 0.2761 | Yes |
| 19 | EHHADH | NA | 2929 | 0.412 | 0.2899 | Yes |
| 20 | SLC27A4 | NA | 3199 | 0.402 | 0.2994 | Yes |
| 21 | ACAD10 | NA | 3267 | 0.399 | 0.3125 | Yes |
| 22 | TYSND1 | NA | 3274 | 0.399 | 0.3266 | Yes |
| 23 | HACL1 | NA | 3370 | 0.396 | 0.3391 | Yes |
| 24 | ACADS | NA | 3399 | 0.395 | 0.3527 | Yes |
| 25 | ABCD3 | NA | 3733 | 0.382 | 0.3603 | Yes |
| 26 | LPIN3 | NA | 3734 | 0.382 | 0.3740 | Yes |
| 27 | ACBD5 | NA | 4668 | 0.354 | 0.3696 | Yes |
| 28 | SCP2 | NA | 4749 | 0.352 | 0.3807 | Yes |
| 29 | PCCB | NA | 5060 | 0.344 | 0.3873 | Yes |
| 30 | ACOX1 | NA | 5098 | 0.343 | 0.3990 | Yes |
| 31 | HADH | NA | 5324 | 0.336 | 0.4069 | Yes |
| 32 | ETFA | NA | 5356 | 0.336 | 0.4183 | Yes |
| 33 | ETFB | NA | 5943 | 0.322 | 0.4191 | Yes |
| 34 | PLA2G15 | NA | 6047 | 0.320 | 0.4287 | Yes |
| 35 | ACAT1 | NA | 6111 | 0.318 | 0.4389 | Yes |
| 36 | NUDT7 | NA | 6183 | 0.317 | 0.4490 | Yes |
| 37 | ABHD2 | NA | 6274 | 0.314 | 0.4586 | Yes |
| 38 | ETFBKMT | NA | 6372 | 0.312 | 0.4680 | Yes |
| 39 | IRS1 | NA | 6700 | 0.305 | 0.4729 | Yes |
| 40 | ACOX2 | NA | 6955 | 0.300 | 0.4790 | Yes |
| 41 | CRAT | NA | 6969 | 0.300 | 0.4895 | Yes |
| 42 | IRS2 | NA | 7052 | 0.298 | 0.4987 | Yes |
| 43 | MECR | NA | 7192 | 0.295 | 0.5067 | Yes |
| 44 | ETFDH | NA | 7555 | 0.288 | 0.5104 | Yes |
| 45 | ECH1 | NA | 7889 | 0.281 | 0.5143 | Yes |
| 46 | AUH | NA | 7930 | 0.280 | 0.5236 | Yes |
| 47 | DECR1 | NA | 8300 | 0.274 | 0.5267 | Yes |
| 48 | ABCD1 | NA | 8410 | 0.272 | 0.5344 | Yes |
| 49 | ADIPOQ | NA | 8737 | 0.266 | 0.5379 | Yes |
| 50 | HADHB | NA | 8884 | 0.263 | 0.5447 | Yes |
| 51 | LPIN2 | NA | 9132 | 0.259 | 0.5494 | Yes |
| 52 | ACAA2 | NA | 9231 | 0.257 | 0.5569 | Yes |
| 53 | ACAA1 | NA | 9324 | 0.255 | 0.5643 | Yes |
| 54 | AMACR | NA | 9352 | 0.255 | 0.5729 | Yes |
| 55 | ACADL | NA | 9951 | 0.245 | 0.5708 | Yes |
| 56 | CPT1B | NA | 10043 | 0.244 | 0.5778 | Yes |
| 57 | PCK2 | NA | 10364 | 0.239 | 0.5805 | Yes |
| 58 | ACOX3 | NA | 10503 | 0.236 | 0.5864 | Yes |
| 59 | ACADVL | NA | 10516 | 0.236 | 0.5947 | Yes |
| 60 | GLYATL2 | NA | 10877 | 0.231 | 0.5963 | Yes |
| 61 | SESN2 | NA | 10921 | 0.230 | 0.6038 | Yes |
| 62 | FABP1 | NA | 11383 | 0.223 | 0.6033 | Yes |
| 63 | FAAH | NA | 12313 | 0.211 | 0.5938 | Yes |
| 64 | PHYH | NA | 12503 | 0.208 | 0.5978 | Yes |
| 65 | CEL | NA | 12514 | 0.208 | 0.6051 | Yes |
| 66 | ECHDC1 | NA | 12905 | 0.202 | 0.6051 | Yes |
| 67 | ILVBL | NA | 12916 | 0.202 | 0.6122 | Yes |
| 68 | MCAT | NA | 13040 | 0.200 | 0.6171 | Yes |
| 69 | ECI2 | NA | 13465 | 0.194 | 0.6163 | Yes |
| 70 | MMAA | NA | 13568 | 0.192 | 0.6213 | Yes |
| 71 | ECHS1 | NA | 13864 | 0.189 | 0.6226 | Yes |
| 72 | PLIN5 | NA | 14018 | 0.186 | 0.6265 | Yes |
| 73 | MFSD2A | NA | 15023 | 0.173 | 0.6143 | No |
| 74 | CROT | NA | 15134 | 0.172 | 0.6184 | No |
| 75 | PECR | NA | 15908 | 0.162 | 0.6101 | No |
| 76 | ACACB | NA | 16170 | 0.159 | 0.6110 | No |
| 77 | PEX2 | NA | 16293 | 0.158 | 0.6144 | No |
| 78 | SLC25A17 | NA | 17872 | 0.138 | 0.5904 | No |
| 79 | HAO2 | NA | 18169 | 0.135 | 0.5898 | No |
| 80 | AIG1 | NA | 18275 | 0.133 | 0.5926 | No |
| 81 | LPIN1 | NA | 18498 | 0.130 | 0.5932 | No |
| 82 | CPT1C | NA | 19017 | 0.124 | 0.5881 | No |
| 83 | CNR1 | NA | 19137 | 0.122 | 0.5903 | No |
| 84 | HAO1 | NA | 19558 | 0.117 | 0.5868 | No |
| 85 | ACAD11 | NA | 19969 | 0.112 | 0.5833 | No |
| 86 | MCEE | NA | 20608 | 0.104 | 0.5753 | No |
| 87 | HADHA | NA | 20662 | 0.104 | 0.5780 | No |
| 88 | ACOT8 | NA | 21338 | 0.096 | 0.5691 | No |
| 89 | HSD17B10 | NA | 21987 | 0.088 | 0.5603 | No |
| 90 | PEX7 | NA | 22878 | 0.079 | 0.5468 | No |
| 91 | ECI1 | NA | 26103 | 0.047 | 0.4893 | No |
| 92 | ABHD1 | NA | 26283 | 0.045 | 0.4877 | No |
| 93 | PCK1 | NA | 26802 | 0.041 | 0.4796 | No |
| 94 | PEX13 | NA | 26897 | 0.040 | 0.4793 | No |
| 95 | ACAT2 | NA | 27532 | 0.036 | 0.4690 | No |
| 96 | CPT1A | NA | 27796 | 0.034 | 0.4654 | No |
| 97 | ABCD2 | NA | 30846 | 0.016 | 0.4100 | No |
| 98 | SLC27A2 | NA | 32521 | 0.009 | 0.3796 | No |
| 99 | ACOXL | NA | 33984 | 0.001 | 0.3527 | No |
| 100 | BDH2 | NA | 40047 | -0.012 | 0.2419 | No |
| 101 | ADTRP | NA | 42484 | -0.042 | 0.1987 | No |
| 102 | PPARD | NA | 47058 | -0.108 | 0.1187 | No |
| 103 | TWIST1 | NA | 49789 | -0.162 | 0.0743 | No |
| 104 | ABCD4 | NA | 50744 | -0.183 | 0.0634 | No |
| 105 | ABCB11 | NA | 51458 | -0.201 | 0.0575 | No |